{
"cells": [
{
"cell_type": "markdown",
"id": "0855b561",
"metadata": {},
"source": [
"# Integrate Modern Data Architectures with Generative AI and interact using prompts for querying SQL databases & APIs"
]
},
{
"cell_type": "markdown",
"id": "26105729-b3e3-42d0-a583-8446fff89277",
"metadata": {},
"source": [
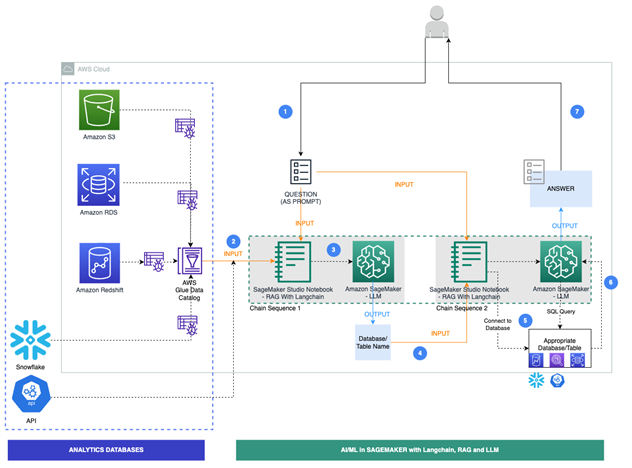
"This notebook demonstrates how **_large language models, such as Flan T5 XL,accessible via SamgeMaker JumpStart_** interact with AWS databases, data stores, and third-party data warehousing solutions like Snowflake. We showcase this interaction 1) by generating and running SQL queries, and 2) making requests to API endpoints. We achieve all of this by using the LangChain framework, which allows the language model to interact with its environment and connect with other sources of data. The LangChain framework operates based on the following principles: calling out to a language model, being data-aware, and being agentic. Our notebook focuses on establishing database connections to various data sources, consolidating metadata, and returning fact-based data points in response to user queries using LLMs and LangChain."
]
},
{
"cell_type": "markdown",
"id": "a310d6ea-2ee1-4979-bb5e-b65cb892c0cd",
"metadata": {},
"source": [
"\n",
" \n"
]
},
{
"cell_type": "markdown",
"id": "5d0297e0-f2dd-464b-9254-6693c45ebafc",
"metadata": {
"tags": []
},
"source": [
"Step 1. Connection to various channels through which LLMs can talk to your data. These channels include:\n",
"\n",
" - RedShift Serverless - to connect to datastore 'tickit'(ticket is referred as tickit in the sample data store) to retrieve information regarding ticket sales.\n",
" - Aurora - MySQL Serverless - to connect to datastore that hosts information about the employees.\n",
" - S3/Athena - to connect to the SageMaker's offline feature store on claims information. \n",
" - Snowflake - to connect to stocks related data residing in finance schema of 3rd party software.\n",
" - APIs - to connect to meteo(in this example we use Langchain's sample dataset on meteo) to retrieve weather information.\n",
" \n",
"Step 2. Usage of Dynamic generation of prompt templates by populating metadata of the tables using Glue Data Catalog(GDC) as context. GDC was populated by running a crawler on the databases. Refer to the information here to create and run a glue crawler. In case of api, a line item was created in GDC data extract.\n",
"\n",
"Step 3. Define Functions to 1/ determine the best data channel to answer the user query, 2/ Generate response to user query\n",
"\n",
"Step 4. Apply user query to LLM and Langchain to determine the data channel. After determining the data channel, run the Langchain SQL Database chain to convert 'text to sql' and run the query against the source data channel. \n",
"\n",
"Finally, display the results.\n"
]
},
{
"cell_type": "markdown",
"id": "e0986ea2-f794-431f-a341-b94f0118cb7d",
"metadata": {
"tags": []
},
"source": [
"### Pre-requisites:\n",
"1. Use kernel Base Python 3.0.\n",
"2. Deploy resources using the cloudformation template mda-llm-cfn.yml.\n",
"\n",
"[OPTIONAL] - If need is to add any of the sources below, then uncomment code in the relevant sections.\n",
"\n",
"1. Setup [Aurora MySQL Serverless database](https://aws.amazon.com/getting-started/hands-on/building-serverless-applications-with-amazon-aurora-serverless/?ref=gsrchandson). Load sample dataset for Human Resource department. Use this notebook to load the data into Aurora MySQL.\n",
"2. Setup [Redshift Serverless](https://catalog.workshops.aws/redshift-immersion/en-US/lab1). Load sample data for Sales & Marketing. For example, 'sample data dev' for 'tickit' dataset available in RedShift examples.\n",
"3. Setup External database. In this case, we are using Snowflake account and populating stocks data. Use this notebook to load the data into Snowflake.\n",
"4. Add/modify the [Glue Crawler](https://catalog.us-east-1.prod.workshops.aws/workshops/71b5bdcf-7eb1-4549-b851-66adc860cd04/en-US/2-studio/1-crawler) on all the databases mentioned above. \n",
"\n",
"**Note - This notebook was tested on kernel - conda_python3 in Region us-east-1**"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "9556eddc-8e45-4e42-9157-213316ec468a",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile requirements.txt\n",
"sqlalchemy==1.4.47\n",
"snowflake-sqlalchemy\n",
"langchain==0.0.166\n",
"sqlalchemy-aurora-data-api\n",
"PyAthena[SQLAlchemy]==2.25.2\n",
"redshift-connector==2.0.910\n",
"sqlalchemy-redshift==0.8.14"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "b55d516c",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"!pip install -r requirements.txt"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "91c153cd",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"import json\n",
"import boto3\n",
"\n",
"import sqlalchemy\n",
"from sqlalchemy import create_engine\n",
"from snowflake.sqlalchemy import URL\n",
"\n",
"from langchain.docstore.document import Document\n",
"from langchain import PromptTemplate,SagemakerEndpoint,SQLDatabase, SQLDatabaseChain, LLMChain\n",
"from langchain.llms.sagemaker_endpoint import LLMContentHandler\n",
"from langchain.chains.question_answering import load_qa_chain\n",
"from langchain.prompts.prompt import PromptTemplate\n",
"from langchain.chains import SQLDatabaseSequentialChain\n",
"\n",
"from langchain.chains.api.prompt import API_RESPONSE_PROMPT\n",
"from langchain.chains import APIChain\n",
"from langchain.prompts.prompt import PromptTemplate\n",
"\n",
"from langchain.chains.api import open_meteo_docs\n",
"\n",
"from typing import Dict"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "f9c445a3-0f6b-4164-83f4-bd5ddf8d084c",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"#define content handler class for flant5xl model\n",
"from langchain.llms.sagemaker_endpoint import LLMContentHandler\n",
"class ContentHandler(LLMContentHandler):\n",
" content_type = \"application/json\"\n",
" accepts = \"application/json\"\n",
" \n",
" def transform_input(self, prompt, model_kwargs) :\n",
" test = {\"text_inputs\": prompt}\n",
" encoded_json = json.dumps(test).encode(\"utf-8\")\n",
" return encoded_json\n",
" \n",
" def transform_output(self, output):\n",
" response_json = json.loads(output.read().decode(\"utf-8\")).get('generated_texts')\n",
" print(\"response\" , response_json)\n",
" return \"\".join(response_json)\n",
"\n",
"content_handler = ContentHandler()"
]
},
{
"cell_type": "markdown",
"id": "074a4144-4053-46c1-ba39-8f64f8fb9e00",
"metadata": {},
"source": [
"The data for this COVID-19 dataset is stored in a public accessible S3 bucket. You can use the following command to explore the dataset.\n",
"\n",
"!aws s3 ls s3://covid19-lake/ --no-sign-request"
]
},
{
"cell_type": "markdown",
"id": "87f05601-529a-42d4-8cab-ba0b9445e695",
"metadata": {
"tags": []
},
"source": [
"### Read parameters from Cloud Formation stack\n",
"Some of the resources needed for this notebook such as the LLM model endpoint, the AWS Glue database and Glue crawler are created through a cloud formation template. The next block of code extracts the outputs and parameters of the cloud formation stack created from that template to get the value of these parameters.\n",
"\n",
"*The stack name here should match the stack name you used when creating the cloud formation stack.*"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "406e3ede-99aa-46e0-b3aa-eb8b65f6cff5",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# if used a different name while creating the cloud formation stack then change this to match the name you used\n",
"CFN_STACK_NAME = \"cfn-genai-mda\""
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "86a1efa5-a1ae-4384-b149-4103b70fab56",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"stacks = boto3.client('cloudformation').list_stacks()\n",
"stack_found = CFN_STACK_NAME in [stack['StackName'] for stack in stacks['StackSummaries']]"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "c0bda7a4-f2a9-4379-b4a9-8e3e17240d0f",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from typing import List\n",
"def get_cfn_outputs(stackname: str) -> List:\n",
" cfn = boto3.client('cloudformation')\n",
" outputs = {}\n",
" for output in cfn.describe_stacks(StackName=stackname)['Stacks'][0]['Outputs']:\n",
" outputs[output['OutputKey']] = output['OutputValue']\n",
" return outputs\n",
"\n",
"def get_cfn_parameters(stackname: str) -> List:\n",
" cfn = boto3.client('cloudformation')\n",
" params = {}\n",
" for param in cfn.describe_stacks(StackName=stackname)['Stacks'][0]['Parameters']:\n",
" params[param['ParameterKey']] = param['ParameterValue']\n",
" return params\n",
"\n",
"if stack_found is True:\n",
" outputs = get_cfn_outputs(CFN_STACK_NAME)\n",
" params = get_cfn_parameters(CFN_STACK_NAME)\n",
" LLMEndpointName = outputs['LLMEndpointName']\n",
" glue_crawler_name = params['CFNCrawlerName']\n",
" glue_database_name = params['CFNDatabaseName']\n",
" glue_databucket_name = params['DataBucketName']\n",
" region = outputs['Region']\n",
" print(f\"cfn outputs={outputs}\\nparams={params}\")\n",
"else:\n",
" print(\"Recheck our cloudformation stack name\")"
]
},
{
"cell_type": "markdown",
"id": "59c5c0b3-d32e-41ff-85fe-dc80586fa737",
"metadata": {
"tags": []
},
"source": [
"### Copy the sample dataset to your S3 bucket"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "ceedd61d-9c21-45b4-b35e-69e5cd047f11",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!aws s3 cp --recursive s3://covid19-lake/rearc-covid-19-testing-data/json/states_daily/ s3://{glue_databucket_name}/covid-dataset/"
]
},
{
"cell_type": "markdown",
"id": "7dbd2b73-cec1-4dbf-8a94-692eb04e7979",
"metadata": {
"tags": []
},
"source": [
"### Run the crawler"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "5ccbd062-0918-4dcf-a4c2-45bfa81e56c7",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile python_glueworkshop.py\n",
"import boto3\n",
"import argparse\n",
"import time\n",
"\n",
"argParser = argparse.ArgumentParser()\n",
"argParser.add_argument(\"-c\", \"--glue_crawler_name\", help=\"script help\")\n",
"args = argParser.parse_args()\n",
"print(args.glue_crawler_name )\n",
"client = boto3.client('glue')\n",
"crawler_name=args.glue_crawler_name\n",
"\n",
"def get_crawler_status(crawler_name):\n",
" # Create a Glue client\n",
" glue_client = boto3.client('glue')\n",
"\n",
" # Get the crawler details\n",
" response = glue_client.get_crawler(Name=crawler_name)\n",
"\n",
" # Extract the crawler state\n",
" crawler_state = response['Crawler']['State']\n",
"\n",
" return crawler_state\n",
"\n",
"# This is the command to start the Crawler\n",
"try:\n",
" response = client.start_crawler(Name=crawler_name )\n",
" print(\"Successfully started crawler. The crawler may take 2-5 mins to detect the schema.\")\n",
"\n",
" while True:\n",
" # Get the crawler status\n",
" status = get_crawler_status(crawler_name)\n",
"\n",
" # Print the crawler status\n",
" print(f\"Crawler '{crawler_name}' status: {status}\")\n",
"\n",
" if status == 'READY': # Replace 'READY' with the desired completed state\n",
" break # Exit the loop if the desired state is reached\n",
"\n",
" time.sleep(10) # Sleep for 10 seconds before checking the status again\n",
" \n",
"except:\n",
" print(\"error in starting crawler. Check the logs for the error details.\")\n"
]
},
{
"cell_type": "markdown",
"id": "d1392725-610f-43c1-8fd9-b5f52c4585b1",
"metadata": {},
"source": [
"Execute the python script by passing the glue crawler name from the cloudformation stack output."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "75d0fa47-1651-4cff-802a-22ae6438a09c",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!python python_glueworkshop.py -c {glue_crawler_name}"
]
},
{
"cell_type": "markdown",
"id": "4132ffc3-6947-49b6-b627-fae3df870b88",
"metadata": {
"tags": []
},
"source": [
"Before proceeding to the next step, check the status of the crawler. It should change from RUNNING to READY. "
]
},
{
"cell_type": "markdown",
"id": "b51d1d0e-33fb-46ca-b82f-6294ea867cae",
"metadata": {
"tags": []
},
"source": [
"### Step 1 - Connect to databases using SQL Alchemy. \n",
"\n",
"Under the hood, LangChain uses SQLAlchemy to connect to SQL databases. The SQLDatabaseChain can therefore be used with any SQL dialect supported by SQLAlchemy, \n",
"such as MS SQL, MySQL, MariaDB, PostgreSQL, Oracle SQL, and SQLite. Please refer to the SQLAlchemy documentation for more information about requirements for connecting to your database. \n"
]
},
{
"cell_type": "markdown",
"id": "e5f5ce28-9b33-4061-8655-2b297d5c24a2",
"metadata": {
"tags": []
},
"source": [
"**Important**: The code below establishes a database connection for data sources and Large Language Models. Please note that the solution will only work if the database connection for your sources is defined in the cell below. Please refer to the Pre-requisites section. If your use case requires data from Aurora MySQL alone, then please comment out other data sources. Furthermore, please update the cluster details and variables for Aurora MySQL accordingly."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "1583cade",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"#define connections\n",
"\n",
"#LLM \n",
"llm=SagemakerEndpoint(\n",
" endpoint_name=LLMEndpointName, \n",
" region_name=region,\n",
" model_kwargs={\"temperature\":1e-10},\n",
" content_handler=content_handler\n",
" )\n",
"\n",
"#S3\n",
"# connect to s3 using athena\n",
"## athena variables\n",
"connathena=f\"athena.{region}.amazonaws.com\" \n",
"portathena='443' #Update, if port is different\n",
"schemaathena=glue_database_name #from cfn params\n",
"s3stagingathena=f's3://{glue_databucket_name}/athenaresults/'#from cfn params\n",
"wkgrpathena='primary'#Update, if workgroup is different\n",
"# tablesathena=['dataset']#[]\n",
"## Create the athena connection string\n",
"connection_string = f\"awsathena+rest://@{connathena}:{portathena}/{schemaathena}?s3_staging_dir={s3stagingathena}/&work_group={wkgrpathena}\"\n",
"## Create the athena SQLAlchemy engine\n",
"engine_athena = create_engine(connection_string, echo=False)\n",
"dbathena = SQLDatabase(engine_athena)\n",
"# dbathena = SQLDatabase(engine_athena, include_tables=tablesathena)\n",
"\n",
"# collect credentials from Secrets Manager\n",
"#Refer here on how to use AWS Secrets Manager - https://docs.aws.amazon.com/secretsmanager/latest/userguide/intro.html\n",
"# client = boto3.client('secretsmanager')\n",
"\n",
"# #SNOWFLAKE\n",
"# # connect to snowflake database\n",
"# ## snowflake variables\n",
"# sf_account_id = \n",
"# sf_secret_id =\n",
"# dwh = \n",
"# db = \n",
"# schema = \n",
"# table =
\n"
]
},
{
"cell_type": "markdown",
"id": "5d0297e0-f2dd-464b-9254-6693c45ebafc",
"metadata": {
"tags": []
},
"source": [
"Step 1. Connection to various channels through which LLMs can talk to your data. These channels include:\n",
"\n",
" - RedShift Serverless - to connect to datastore 'tickit'(ticket is referred as tickit in the sample data store) to retrieve information regarding ticket sales.\n",
" - Aurora - MySQL Serverless - to connect to datastore that hosts information about the employees.\n",
" - S3/Athena - to connect to the SageMaker's offline feature store on claims information. \n",
" - Snowflake - to connect to stocks related data residing in finance schema of 3rd party software.\n",
" - APIs - to connect to meteo(in this example we use Langchain's sample dataset on meteo) to retrieve weather information.\n",
" \n",
"Step 2. Usage of Dynamic generation of prompt templates by populating metadata of the tables using Glue Data Catalog(GDC) as context. GDC was populated by running a crawler on the databases. Refer to the information here to create and run a glue crawler. In case of api, a line item was created in GDC data extract.\n",
"\n",
"Step 3. Define Functions to 1/ determine the best data channel to answer the user query, 2/ Generate response to user query\n",
"\n",
"Step 4. Apply user query to LLM and Langchain to determine the data channel. After determining the data channel, run the Langchain SQL Database chain to convert 'text to sql' and run the query against the source data channel. \n",
"\n",
"Finally, display the results.\n"
]
},
{
"cell_type": "markdown",
"id": "e0986ea2-f794-431f-a341-b94f0118cb7d",
"metadata": {
"tags": []
},
"source": [
"### Pre-requisites:\n",
"1. Use kernel Base Python 3.0.\n",
"2. Deploy resources using the cloudformation template mda-llm-cfn.yml.\n",
"\n",
"[OPTIONAL] - If need is to add any of the sources below, then uncomment code in the relevant sections.\n",
"\n",
"1. Setup [Aurora MySQL Serverless database](https://aws.amazon.com/getting-started/hands-on/building-serverless-applications-with-amazon-aurora-serverless/?ref=gsrchandson). Load sample dataset for Human Resource department. Use this notebook to load the data into Aurora MySQL.\n",
"2. Setup [Redshift Serverless](https://catalog.workshops.aws/redshift-immersion/en-US/lab1). Load sample data for Sales & Marketing. For example, 'sample data dev' for 'tickit' dataset available in RedShift examples.\n",
"3. Setup External database. In this case, we are using Snowflake account and populating stocks data. Use this notebook to load the data into Snowflake.\n",
"4. Add/modify the [Glue Crawler](https://catalog.us-east-1.prod.workshops.aws/workshops/71b5bdcf-7eb1-4549-b851-66adc860cd04/en-US/2-studio/1-crawler) on all the databases mentioned above. \n",
"\n",
"**Note - This notebook was tested on kernel - conda_python3 in Region us-east-1**"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "9556eddc-8e45-4e42-9157-213316ec468a",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile requirements.txt\n",
"sqlalchemy==1.4.47\n",
"snowflake-sqlalchemy\n",
"langchain==0.0.166\n",
"sqlalchemy-aurora-data-api\n",
"PyAthena[SQLAlchemy]==2.25.2\n",
"redshift-connector==2.0.910\n",
"sqlalchemy-redshift==0.8.14"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "b55d516c",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"!pip install -r requirements.txt"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "91c153cd",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"import json\n",
"import boto3\n",
"\n",
"import sqlalchemy\n",
"from sqlalchemy import create_engine\n",
"from snowflake.sqlalchemy import URL\n",
"\n",
"from langchain.docstore.document import Document\n",
"from langchain import PromptTemplate,SagemakerEndpoint,SQLDatabase, SQLDatabaseChain, LLMChain\n",
"from langchain.llms.sagemaker_endpoint import LLMContentHandler\n",
"from langchain.chains.question_answering import load_qa_chain\n",
"from langchain.prompts.prompt import PromptTemplate\n",
"from langchain.chains import SQLDatabaseSequentialChain\n",
"\n",
"from langchain.chains.api.prompt import API_RESPONSE_PROMPT\n",
"from langchain.chains import APIChain\n",
"from langchain.prompts.prompt import PromptTemplate\n",
"\n",
"from langchain.chains.api import open_meteo_docs\n",
"\n",
"from typing import Dict"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "f9c445a3-0f6b-4164-83f4-bd5ddf8d084c",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"#define content handler class for flant5xl model\n",
"from langchain.llms.sagemaker_endpoint import LLMContentHandler\n",
"class ContentHandler(LLMContentHandler):\n",
" content_type = \"application/json\"\n",
" accepts = \"application/json\"\n",
" \n",
" def transform_input(self, prompt, model_kwargs) :\n",
" test = {\"text_inputs\": prompt}\n",
" encoded_json = json.dumps(test).encode(\"utf-8\")\n",
" return encoded_json\n",
" \n",
" def transform_output(self, output):\n",
" response_json = json.loads(output.read().decode(\"utf-8\")).get('generated_texts')\n",
" print(\"response\" , response_json)\n",
" return \"\".join(response_json)\n",
"\n",
"content_handler = ContentHandler()"
]
},
{
"cell_type": "markdown",
"id": "074a4144-4053-46c1-ba39-8f64f8fb9e00",
"metadata": {},
"source": [
"The data for this COVID-19 dataset is stored in a public accessible S3 bucket. You can use the following command to explore the dataset.\n",
"\n",
"!aws s3 ls s3://covid19-lake/ --no-sign-request"
]
},
{
"cell_type": "markdown",
"id": "87f05601-529a-42d4-8cab-ba0b9445e695",
"metadata": {
"tags": []
},
"source": [
"### Read parameters from Cloud Formation stack\n",
"Some of the resources needed for this notebook such as the LLM model endpoint, the AWS Glue database and Glue crawler are created through a cloud formation template. The next block of code extracts the outputs and parameters of the cloud formation stack created from that template to get the value of these parameters.\n",
"\n",
"*The stack name here should match the stack name you used when creating the cloud formation stack.*"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "406e3ede-99aa-46e0-b3aa-eb8b65f6cff5",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# if used a different name while creating the cloud formation stack then change this to match the name you used\n",
"CFN_STACK_NAME = \"cfn-genai-mda\""
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "86a1efa5-a1ae-4384-b149-4103b70fab56",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"stacks = boto3.client('cloudformation').list_stacks()\n",
"stack_found = CFN_STACK_NAME in [stack['StackName'] for stack in stacks['StackSummaries']]"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "c0bda7a4-f2a9-4379-b4a9-8e3e17240d0f",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from typing import List\n",
"def get_cfn_outputs(stackname: str) -> List:\n",
" cfn = boto3.client('cloudformation')\n",
" outputs = {}\n",
" for output in cfn.describe_stacks(StackName=stackname)['Stacks'][0]['Outputs']:\n",
" outputs[output['OutputKey']] = output['OutputValue']\n",
" return outputs\n",
"\n",
"def get_cfn_parameters(stackname: str) -> List:\n",
" cfn = boto3.client('cloudformation')\n",
" params = {}\n",
" for param in cfn.describe_stacks(StackName=stackname)['Stacks'][0]['Parameters']:\n",
" params[param['ParameterKey']] = param['ParameterValue']\n",
" return params\n",
"\n",
"if stack_found is True:\n",
" outputs = get_cfn_outputs(CFN_STACK_NAME)\n",
" params = get_cfn_parameters(CFN_STACK_NAME)\n",
" LLMEndpointName = outputs['LLMEndpointName']\n",
" glue_crawler_name = params['CFNCrawlerName']\n",
" glue_database_name = params['CFNDatabaseName']\n",
" glue_databucket_name = params['DataBucketName']\n",
" region = outputs['Region']\n",
" print(f\"cfn outputs={outputs}\\nparams={params}\")\n",
"else:\n",
" print(\"Recheck our cloudformation stack name\")"
]
},
{
"cell_type": "markdown",
"id": "59c5c0b3-d32e-41ff-85fe-dc80586fa737",
"metadata": {
"tags": []
},
"source": [
"### Copy the sample dataset to your S3 bucket"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "ceedd61d-9c21-45b4-b35e-69e5cd047f11",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!aws s3 cp --recursive s3://covid19-lake/rearc-covid-19-testing-data/json/states_daily/ s3://{glue_databucket_name}/covid-dataset/"
]
},
{
"cell_type": "markdown",
"id": "7dbd2b73-cec1-4dbf-8a94-692eb04e7979",
"metadata": {
"tags": []
},
"source": [
"### Run the crawler"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "5ccbd062-0918-4dcf-a4c2-45bfa81e56c7",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile python_glueworkshop.py\n",
"import boto3\n",
"import argparse\n",
"import time\n",
"\n",
"argParser = argparse.ArgumentParser()\n",
"argParser.add_argument(\"-c\", \"--glue_crawler_name\", help=\"script help\")\n",
"args = argParser.parse_args()\n",
"print(args.glue_crawler_name )\n",
"client = boto3.client('glue')\n",
"crawler_name=args.glue_crawler_name\n",
"\n",
"def get_crawler_status(crawler_name):\n",
" # Create a Glue client\n",
" glue_client = boto3.client('glue')\n",
"\n",
" # Get the crawler details\n",
" response = glue_client.get_crawler(Name=crawler_name)\n",
"\n",
" # Extract the crawler state\n",
" crawler_state = response['Crawler']['State']\n",
"\n",
" return crawler_state\n",
"\n",
"# This is the command to start the Crawler\n",
"try:\n",
" response = client.start_crawler(Name=crawler_name )\n",
" print(\"Successfully started crawler. The crawler may take 2-5 mins to detect the schema.\")\n",
"\n",
" while True:\n",
" # Get the crawler status\n",
" status = get_crawler_status(crawler_name)\n",
"\n",
" # Print the crawler status\n",
" print(f\"Crawler '{crawler_name}' status: {status}\")\n",
"\n",
" if status == 'READY': # Replace 'READY' with the desired completed state\n",
" break # Exit the loop if the desired state is reached\n",
"\n",
" time.sleep(10) # Sleep for 10 seconds before checking the status again\n",
" \n",
"except:\n",
" print(\"error in starting crawler. Check the logs for the error details.\")\n"
]
},
{
"cell_type": "markdown",
"id": "d1392725-610f-43c1-8fd9-b5f52c4585b1",
"metadata": {},
"source": [
"Execute the python script by passing the glue crawler name from the cloudformation stack output."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "75d0fa47-1651-4cff-802a-22ae6438a09c",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!python python_glueworkshop.py -c {glue_crawler_name}"
]
},
{
"cell_type": "markdown",
"id": "4132ffc3-6947-49b6-b627-fae3df870b88",
"metadata": {
"tags": []
},
"source": [
"Before proceeding to the next step, check the status of the crawler. It should change from RUNNING to READY. "
]
},
{
"cell_type": "markdown",
"id": "b51d1d0e-33fb-46ca-b82f-6294ea867cae",
"metadata": {
"tags": []
},
"source": [
"### Step 1 - Connect to databases using SQL Alchemy. \n",
"\n",
"Under the hood, LangChain uses SQLAlchemy to connect to SQL databases. The SQLDatabaseChain can therefore be used with any SQL dialect supported by SQLAlchemy, \n",
"such as MS SQL, MySQL, MariaDB, PostgreSQL, Oracle SQL, and SQLite. Please refer to the SQLAlchemy documentation for more information about requirements for connecting to your database. \n"
]
},
{
"cell_type": "markdown",
"id": "e5f5ce28-9b33-4061-8655-2b297d5c24a2",
"metadata": {
"tags": []
},
"source": [
"**Important**: The code below establishes a database connection for data sources and Large Language Models. Please note that the solution will only work if the database connection for your sources is defined in the cell below. Please refer to the Pre-requisites section. If your use case requires data from Aurora MySQL alone, then please comment out other data sources. Furthermore, please update the cluster details and variables for Aurora MySQL accordingly."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "1583cade",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"#define connections\n",
"\n",
"#LLM \n",
"llm=SagemakerEndpoint(\n",
" endpoint_name=LLMEndpointName, \n",
" region_name=region,\n",
" model_kwargs={\"temperature\":1e-10},\n",
" content_handler=content_handler\n",
" )\n",
"\n",
"#S3\n",
"# connect to s3 using athena\n",
"## athena variables\n",
"connathena=f\"athena.{region}.amazonaws.com\" \n",
"portathena='443' #Update, if port is different\n",
"schemaathena=glue_database_name #from cfn params\n",
"s3stagingathena=f's3://{glue_databucket_name}/athenaresults/'#from cfn params\n",
"wkgrpathena='primary'#Update, if workgroup is different\n",
"# tablesathena=['dataset']#[]\n",
"## Create the athena connection string\n",
"connection_string = f\"awsathena+rest://@{connathena}:{portathena}/{schemaathena}?s3_staging_dir={s3stagingathena}/&work_group={wkgrpathena}\"\n",
"## Create the athena SQLAlchemy engine\n",
"engine_athena = create_engine(connection_string, echo=False)\n",
"dbathena = SQLDatabase(engine_athena)\n",
"# dbathena = SQLDatabase(engine_athena, include_tables=tablesathena)\n",
"\n",
"# collect credentials from Secrets Manager\n",
"#Refer here on how to use AWS Secrets Manager - https://docs.aws.amazon.com/secretsmanager/latest/userguide/intro.html\n",
"# client = boto3.client('secretsmanager')\n",
"\n",
"# #SNOWFLAKE\n",
"# # connect to snowflake database\n",
"# ## snowflake variables\n",
"# sf_account_id = \n",
"# sf_secret_id =\n",
"# dwh = \n",
"# db = \n",
"# schema = \n",
"# table = \n",
"# ## snowflake get credentials from secrets manager\n",
"# response = client.get_secret_value(SecretId=sf_secret_id)\n",
"# secrets_credentials = json.loads(response['SecretString'])\n",
"# sf_password = secrets_credentials['password']\n",
"# sf_username = secrets_credentials['username']\n",
"# ## Create the snowflake connection string\n",
"# connection_string = f\"snowflake://{sf_username}:{sf_password}@{sf_account_id}/{db}/{schema}?warehouse={dwh}\"\n",
"# ## Create the snowflake SQLAlchemy engine\n",
"# engine_snowflake = create_engine(connection_string, echo=False)\n",
"# dbsnowflake = SQLDatabase(engine_snowflake)\n",
"\n",
"# #AURORA MYSQL\n",
"# ##connect to aurora mysql\n",
"# ##aurora mysql cluster details/variables\n",
"# cluster_arn = \n",
"# secret_arn =\n",
"# rdsdb=\n",
"# rdsdb_tbl = []\n",
"# ## Create the aurora connection string\n",
"# connection_string = f\"mysql+auroradataapi://:@/{rdsdb}\"\n",
"# ## Create the aurora SQLAlchemy engine\n",
"# engine_rds = create_engine(connection_string, echo=False,connect_args=dict(aurora_cluster_arn=cluster_arn, secret_arn=secret_arn))\n",
"# dbrds = SQLDatabase(engine_rds, include_tables=rdsdb_tbl)\n",
"\n",
"# #REDSHIFT\n",
"# # connect to redshift database\n",
"# ## redshift variables\n",
"# rs_secret_id = \n",
"# rs_endpoint=\n",
"# rs_port=\n",
"# rs_db=\n",
"# rs_schema=\n",
"# ## redshift get credentials from secrets manager\n",
"# response = client.get_secret_value(SecretId=rs_secret_id)\n",
"# secrets_credentials = json.loads(response['SecretString'])\n",
"# rs_password = secrets_credentials['password']\n",
"# rs_username = secrets_credentials['username']\n",
"# ## Create the redshift connection string\n",
"# connection_string = f\"redshift+redshift_connector://{rs_username}:{rs_password}@{rs_endpoint}:{rs_port}/{rs_db}\"\n",
"# engine_redshift = create_engine(connection_string, echo=False)\n",
"# dbredshift = SQLDatabase(engine_redshift)\n",
"\n",
"#Glue Data Catalog\n",
"##Provide list of all the databases where the table metadata resides after the glue successfully crawls the table\n",
"# gdc = ['redshift-sagemaker-sample-data-dev', 'snowflake','rds-aurora-mysql-employees','sagemaker_featurestore'] # mentioned a few examples here\n",
"gdc = [schemaathena] \n"
]
},
{
"cell_type": "markdown",
"id": "1ea21757-b08a-438b-a5a7-79d85a9a9085",
"metadata": {},
"source": [
"### Step 2 - Generate Dynamic Prompt Templates\n",
"Build a consolidated view of Glue Data Catalog by combining metadata stored for all the databases in pipe delimited format."
]
},
{
"cell_type": "code",
"execution_count": 12,
"id": "08a3373d-9285-4fab-81b5-51e5364590b5",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"s3|cfn_covid_lake|cfn_covid_dataset|totaltestresults\n",
"s3|cfn_covid_lake|cfn_covid_dataset|fips\n",
"s3|cfn_covid_lake|cfn_covid_dataset|deathincrease\n",
"s3|cfn_covid_lake|cfn_covid_dataset|hospitalizedincrease\n",
"s3|cfn_covid_lake|cfn_covid_dataset|negativeincrease\n",
"s3|cfn_covid_lake|cfn_covid_dataset|positiveincrease\n",
"s3|cfn_covid_lake|cfn_covid_dataset|totaltestresultsincrease\n",
"s3|cfn_covid_lake|cfn_covid_dataset|negative\n",
"s3|cfn_covid_lake|cfn_covid_dataset|pending\n",
"api|meteo|weather|weather\n"
]
}
],
"source": [
"#Generate Dynamic prompts to populate the Glue Data Catalog\n",
"#harvest aws crawler metadata\n",
"\n",
"def parse_catalog():\n",
" #Connect to Glue catalog\n",
" #get metadata of redshift serverless tables\n",
" columns_str=''\n",
" \n",
" #define glue cient\n",
" glue_client = boto3.client('glue')\n",
" \n",
" for db in gdc:\n",
" response = glue_client.get_tables(DatabaseName =db)\n",
" for tables in response['TableList']:\n",
" #classification in the response for s3 and other databases is different. Set classification based on the response location\n",
" if tables['StorageDescriptor']['Location'].startswith('s3'): classification='s3' \n",
" else: classification = tables['Parameters']['classification']\n",
" for columns in tables['StorageDescriptor']['Columns']:\n",
" dbname,tblname,colname=tables['DatabaseName'],tables['Name'],columns['Name']\n",
" columns_str=columns_str+f'\\n{classification}|{dbname}|{tblname}|{colname}' \n",
" #API\n",
" ## Append the metadata of the API to the unified glue data catalog\n",
" columns_str=columns_str+'\\n'+('api|meteo|weather|weather')\n",
" return columns_str\n",
"\n",
"glue_catalog = parse_catalog()\n",
"\n",
"#display a few lines from the catalog\n",
"print('\\n'.join(glue_catalog.splitlines()[-10:]) )\n"
]
},
{
"cell_type": "markdown",
"id": "a94e6770-42c3-402b-a60e-9c21fb99d5f6",
"metadata": {
"tags": []
},
"source": [
"### Step 3 - Define Functions to 1/ determine the best data channel to answer the user query, 2/ Generate response to user query"
]
},
{
"cell_type": "markdown",
"id": "adda3714-3f32-4480-9526-91cca37489d1",
"metadata": {},
"source": [
"In this code sample, we use the Anthropic Model to generate inferences. You can utilize SageMaker JumpStart models to achieve the same. \n",
"Guidance on how to use the JumpStart Models is available in the notebook - mda_with_llm_langchain_smjumpstart_flant5xl"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "4efcc59b",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"#Function 1 'Infer Channel'\n",
"#define a function that infers the channel/database/table and sets the database for querying\n",
"def identify_channel(query):\n",
" #Prompt 1 'Infer Channel'\n",
" ##set prompt template. It instructs the llm on how to evaluate and respond to the llm. It is referred to as dynamic since glue data catalog is first getting generated and appended to the prompt.\n",
" prompt_template = \"\"\"\n",
" From the table below, find the database (in column database) which will contain the data (in corresponding column_names) to answer the question \n",
" {query} \\n\n",
" \"\"\"+glue_catalog +\"\"\" \n",
" Give your answer as database == \n",
" Also,give your answer as database.table == \n",
" \"\"\"\n",
" ##define prompt 1\n",
" PROMPT_channel = PromptTemplate( template=prompt_template, input_variables=[\"query\"] )\n",
"\n",
" # define llm chain\n",
" llm_chain = LLMChain(prompt=PROMPT_channel, llm=llm)\n",
" #run the query and save to generated texts\n",
" generated_texts = llm_chain.run(query)\n",
" print(generated_texts)\n",
"\n",
" #set the best channel from where the query can be answered\n",
" if 'snowflake' in generated_texts: \n",
" channel='db'\n",
" db=dbsnowflake \n",
" print(\"SET database to snowflake\") \n",
" elif 'redshift' in generated_texts: \n",
" channel='db'\n",
" db=dbredshift\n",
" print(\"SET database to redshift\")\n",
" elif 's3' in generated_texts: \n",
" channel='db'\n",
" db=dbathena\n",
" print(\"SET database to athena\")\n",
" elif 'rdsmysql' in generated_texts: \n",
" channel='db'\n",
" db=dbrds\n",
" print(\"SET database to rds\") \n",
" elif 'api' in generated_texts: \n",
" channel='api'\n",
" print(\"SET database to weather api\") \n",
" else: raise Exception(\"User question cannot be answered by any of the channels mentioned in the catalog\")\n",
" print(\"Step complete. Channel is: \", channel)\n",
" \n",
" return channel, db\n",
"\n",
"#Function 2 'Run Query'\n",
"#define a function that infers the channel/database/table and sets the database for querying\n",
"def run_query(query):\n",
"\n",
" channel, db = identify_channel(query) #call the identify channel function first\n",
"\n",
" ##Prompt 2 'Run Query'\n",
" #after determining the data channel, run the Langchain SQL Database chain to convert 'text to sql' and run the query against the source data channel. \n",
" #provide rules for running the SQL queries in default template--> table info.\n",
"\n",
" _DEFAULT_TEMPLATE = \"\"\"Given an input question, first create a syntactically correct {dialect} query to run, then look at the results of the query and return the answer.\n",
"\n",
" Only use the following tables:\n",
"\n",
" {table_info}\n",
" if someone asks for covid data, then use the table cfn_covid_lake.cfn_covid_dataset.\n",
"\n",
" Question: {input}\"\"\"\n",
"\n",
" PROMPT_sql = PromptTemplate(\n",
" input_variables=[\"input\", \"table_info\", \"dialect\"], template=_DEFAULT_TEMPLATE\n",
" )\n",
"\n",
" \n",
" if channel=='db':\n",
" db_chain = SQLDatabaseChain.from_llm(llm, db, prompt=PROMPT_sql, verbose=False, return_intermediate_steps=False)\n",
" response=db_chain.run(query)\n",
" elif channel=='api':\n",
" chain_api = APIChain.from_llm_and_api_docs(llm, open_meteo_docs.OPEN_METEO_DOCS, verbose=True)\n",
" response=chain_api.run(query)\n",
" else: raise Exception(\"Unlisted channel. Check your unified catalog\")\n",
" return response\n",
"\n"
]
},
{
"cell_type": "markdown",
"id": "390a92cd-e1b4-4feb-ab7a-f97030ba7f84",
"metadata": {},
"source": [
"### Step 4 - Run the run_query function that in turn calls the Langchain SQL Database chain to convert 'text to sql' and runs the query against the source data channel\n",
"\n",
"Some samples are provided below for test runs. Uncomment the query to run."
]
},
{
"cell_type": "code",
"execution_count": 14,
"id": "f82599a2",
"metadata": {
"tags": []
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"response ['s3|cfn_covid_lake|cfn_covid']\n",
"s3|cfn_covid_lake|cfn_covid\n",
"SET database to athena\n",
"Step complete. Channel is: db\n",
"response ['SELECT count(*) FROM cfn_covid_dataset W']\n",
"response ['[103900]']\n",
"----------------------------------------------------------------------\n",
"SQL and response from user query How many covid cases are there? \n",
" [103900]\n"
]
}
],
"source": [
"# Enter the query\n",
"## Few queries to try out - \n",
"#athena - Healthcare - Covid dataset\n",
"# query = \"\"\"How many covid hospitalizations were reported in NY in June of 2021?\"\"\" \n",
"# query = \"\"\"Which States reported the least and maximum deaths?\"\"\" \n",
"query=\"How many covid cases are there?\"\n",
"\n",
"#snowflake - Finance and Investments\n",
"# query = \"\"\"Which stock performed the best and the worst in May of 2013?\"\"\"\n",
"# query = \"\"\"What is the average volume stocks traded in July of 2013?\"\"\"\n",
"\n",
"#rds - Human Resources\n",
"# query = \"\"\"Name all employees with birth date this month\"\"\" \n",
"# query = \"\"\"Combien d'employés sont des femmes? \"\"\" #Ask question in French - How many females are there?\n",
"# query = \"\"\"How many employees were hired before 1990?\"\"\" \n",
"\n",
"#athena - Legal - SageMaker offline featurestore\n",
"# query = \"\"\"How many frauds happened in the year 2023 ?\"\"\" \n",
"# query = \"\"\"How many policies were claimed this year ?\"\"\" \n",
"\n",
"#redshift - Sales & Marketing\n",
"# query = \"\"\"How many tickit sales are there\"\"\" \n",
"# query = \"what was the total commision for the tickit sales in the year 2008?\" \n",
"\n",
"#api - product - weather\n",
"# query = \"\"\"What is the weather like right now in New York City in degrees Farenheit?\"\"\"\n",
"\n",
"#Response from Langchain\n",
"response = run_query(query)\n",
"print(\"----------------------------------------------------------------------\")\n",
"print(f'SQL and response from user query {query} \\n {response}')"
]
},
{
"cell_type": "markdown",
"id": "69371bdc-537f-4e5e-a004-99d852097862",
"metadata": {},
"source": [
"### Clean-up\n",
"After you run the modern data architecture with Generative AI, make sure to clean up any resources that won’t be utilized. Shutdown and delete the databases used (Amazon Redshift, Amazon RDS, Snowflake). In addition, delete the data in Amazon S3 and make sure to stop any SageMaker Studio notebook instances to not incur any further charges. If you used SageMaker Jumpstart to deploy large language model as SageMaker Real-time Endpoint, delete endpoint either through SageMaker console, or through Studio. \n",
"\n",
"To completely remove all the provisoned resources, go to CloudFormation and delete the stack.\n"
]
}
],
"metadata": {
"availableInstances": [
{
"_defaultOrder": 0,
"_isFastLaunch": true,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 4,
"name": "ml.t3.medium",
"vcpuNum": 2
},
{
"_defaultOrder": 1,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 8,
"name": "ml.t3.large",
"vcpuNum": 2
},
{
"_defaultOrder": 2,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 16,
"name": "ml.t3.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 3,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 32,
"name": "ml.t3.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 4,
"_isFastLaunch": true,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 8,
"name": "ml.m5.large",
"vcpuNum": 2
},
{
"_defaultOrder": 5,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 16,
"name": "ml.m5.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 6,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 32,
"name": "ml.m5.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 7,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 64,
"name": "ml.m5.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 8,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 128,
"name": "ml.m5.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 9,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 192,
"name": "ml.m5.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 10,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 256,
"name": "ml.m5.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 11,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 384,
"name": "ml.m5.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 12,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 8,
"name": "ml.m5d.large",
"vcpuNum": 2
},
{
"_defaultOrder": 13,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 16,
"name": "ml.m5d.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 14,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 32,
"name": "ml.m5d.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 15,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 64,
"name": "ml.m5d.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 16,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 128,
"name": "ml.m5d.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 17,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 192,
"name": "ml.m5d.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 18,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 256,
"name": "ml.m5d.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 19,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 384,
"name": "ml.m5d.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 20,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"hideHardwareSpecs": true,
"memoryGiB": 0,
"name": "ml.geospatial.interactive",
"supportedImageNames": [
"sagemaker-geospatial-v1-0"
],
"vcpuNum": 0
},
{
"_defaultOrder": 21,
"_isFastLaunch": true,
"category": "Compute optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 4,
"name": "ml.c5.large",
"vcpuNum": 2
},
{
"_defaultOrder": 22,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 8,
"name": "ml.c5.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 23,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 16,
"name": "ml.c5.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 24,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 32,
"name": "ml.c5.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 25,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 72,
"name": "ml.c5.9xlarge",
"vcpuNum": 36
},
{
"_defaultOrder": 26,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 96,
"name": "ml.c5.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 27,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 144,
"name": "ml.c5.18xlarge",
"vcpuNum": 72
},
{
"_defaultOrder": 28,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 192,
"name": "ml.c5.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 29,
"_isFastLaunch": true,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 16,
"name": "ml.g4dn.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 30,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 32,
"name": "ml.g4dn.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 31,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 64,
"name": "ml.g4dn.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 32,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 128,
"name": "ml.g4dn.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 33,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 4,
"hideHardwareSpecs": false,
"memoryGiB": 192,
"name": "ml.g4dn.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 34,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 256,
"name": "ml.g4dn.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 35,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 61,
"name": "ml.p3.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 36,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 4,
"hideHardwareSpecs": false,
"memoryGiB": 244,
"name": "ml.p3.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 37,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 8,
"hideHardwareSpecs": false,
"memoryGiB": 488,
"name": "ml.p3.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 38,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 8,
"hideHardwareSpecs": false,
"memoryGiB": 768,
"name": "ml.p3dn.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 39,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 16,
"name": "ml.r5.large",
"vcpuNum": 2
},
{
"_defaultOrder": 40,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 32,
"name": "ml.r5.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 41,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 64,
"name": "ml.r5.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 42,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 128,
"name": "ml.r5.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 43,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 256,
"name": "ml.r5.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 44,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 384,
"name": "ml.r5.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 45,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 512,
"name": "ml.r5.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 46,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"hideHardwareSpecs": false,
"memoryGiB": 768,
"name": "ml.r5.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 47,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 16,
"name": "ml.g5.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 48,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 32,
"name": "ml.g5.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 49,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 64,
"name": "ml.g5.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 50,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 128,
"name": "ml.g5.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 51,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"hideHardwareSpecs": false,
"memoryGiB": 256,
"name": "ml.g5.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 52,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 4,
"hideHardwareSpecs": false,
"memoryGiB": 192,
"name": "ml.g5.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 53,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 4,
"hideHardwareSpecs": false,
"memoryGiB": 384,
"name": "ml.g5.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 54,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 8,
"hideHardwareSpecs": false,
"memoryGiB": 768,
"name": "ml.g5.48xlarge",
"vcpuNum": 192
}
],
"instance_type": "ml.t3.medium",
"kernelspec": {
"display_name": "conda_python3",
"language": "python",
"name": "conda_python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.10.8"
}
},
"nbformat": 4,
"nbformat_minor": 5
}
 \n"
]
},
{
"cell_type": "markdown",
"id": "5d0297e0-f2dd-464b-9254-6693c45ebafc",
"metadata": {
"tags": []
},
"source": [
"Step 1. Connection to various channels through which LLMs can talk to your data. These channels include:\n",
"\n",
" - RedShift Serverless - to connect to datastore 'tickit'(ticket is referred as tickit in the sample data store) to retrieve information regarding ticket sales.\n",
" - Aurora - MySQL Serverless - to connect to datastore that hosts information about the employees.\n",
" - S3/Athena - to connect to the SageMaker's offline feature store on claims information. \n",
" - Snowflake - to connect to stocks related data residing in finance schema of 3rd party software.\n",
" - APIs - to connect to meteo(in this example we use Langchain's sample dataset on meteo) to retrieve weather information.\n",
" \n",
"Step 2. Usage of Dynamic generation of prompt templates by populating metadata of the tables using Glue Data Catalog(GDC) as context. GDC was populated by running a crawler on the databases. Refer to the information here to create and run a glue crawler. In case of api, a line item was created in GDC data extract.\n",
"\n",
"Step 3. Define Functions to 1/ determine the best data channel to answer the user query, 2/ Generate response to user query\n",
"\n",
"Step 4. Apply user query to LLM and Langchain to determine the data channel. After determining the data channel, run the Langchain SQL Database chain to convert 'text to sql' and run the query against the source data channel. \n",
"\n",
"Finally, display the results.\n"
]
},
{
"cell_type": "markdown",
"id": "e0986ea2-f794-431f-a341-b94f0118cb7d",
"metadata": {
"tags": []
},
"source": [
"### Pre-requisites:\n",
"1. Use kernel Base Python 3.0.\n",
"2. Deploy resources using the cloudformation template mda-llm-cfn.yml.\n",
"\n",
"[OPTIONAL] - If need is to add any of the sources below, then uncomment code in the relevant sections.\n",
"\n",
"1. Setup [Aurora MySQL Serverless database](https://aws.amazon.com/getting-started/hands-on/building-serverless-applications-with-amazon-aurora-serverless/?ref=gsrchandson). Load sample dataset for Human Resource department. Use this notebook to load the data into Aurora MySQL.\n",
"2. Setup [Redshift Serverless](https://catalog.workshops.aws/redshift-immersion/en-US/lab1). Load sample data for Sales & Marketing. For example, 'sample data dev' for 'tickit' dataset available in RedShift examples.\n",
"3. Setup External database. In this case, we are using Snowflake account and populating stocks data. Use this notebook to load the data into Snowflake.\n",
"4. Add/modify the [Glue Crawler](https://catalog.us-east-1.prod.workshops.aws/workshops/71b5bdcf-7eb1-4549-b851-66adc860cd04/en-US/2-studio/1-crawler) on all the databases mentioned above. \n",
"\n",
"**Note - This notebook was tested on kernel - conda_python3 in Region us-east-1**"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "9556eddc-8e45-4e42-9157-213316ec468a",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile requirements.txt\n",
"sqlalchemy==1.4.47\n",
"snowflake-sqlalchemy\n",
"langchain==0.0.166\n",
"sqlalchemy-aurora-data-api\n",
"PyAthena[SQLAlchemy]==2.25.2\n",
"redshift-connector==2.0.910\n",
"sqlalchemy-redshift==0.8.14"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "b55d516c",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"!pip install -r requirements.txt"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "91c153cd",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"import json\n",
"import boto3\n",
"\n",
"import sqlalchemy\n",
"from sqlalchemy import create_engine\n",
"from snowflake.sqlalchemy import URL\n",
"\n",
"from langchain.docstore.document import Document\n",
"from langchain import PromptTemplate,SagemakerEndpoint,SQLDatabase, SQLDatabaseChain, LLMChain\n",
"from langchain.llms.sagemaker_endpoint import LLMContentHandler\n",
"from langchain.chains.question_answering import load_qa_chain\n",
"from langchain.prompts.prompt import PromptTemplate\n",
"from langchain.chains import SQLDatabaseSequentialChain\n",
"\n",
"from langchain.chains.api.prompt import API_RESPONSE_PROMPT\n",
"from langchain.chains import APIChain\n",
"from langchain.prompts.prompt import PromptTemplate\n",
"\n",
"from langchain.chains.api import open_meteo_docs\n",
"\n",
"from typing import Dict"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "f9c445a3-0f6b-4164-83f4-bd5ddf8d084c",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"#define content handler class for flant5xl model\n",
"from langchain.llms.sagemaker_endpoint import LLMContentHandler\n",
"class ContentHandler(LLMContentHandler):\n",
" content_type = \"application/json\"\n",
" accepts = \"application/json\"\n",
" \n",
" def transform_input(self, prompt, model_kwargs) :\n",
" test = {\"text_inputs\": prompt}\n",
" encoded_json = json.dumps(test).encode(\"utf-8\")\n",
" return encoded_json\n",
" \n",
" def transform_output(self, output):\n",
" response_json = json.loads(output.read().decode(\"utf-8\")).get('generated_texts')\n",
" print(\"response\" , response_json)\n",
" return \"\".join(response_json)\n",
"\n",
"content_handler = ContentHandler()"
]
},
{
"cell_type": "markdown",
"id": "074a4144-4053-46c1-ba39-8f64f8fb9e00",
"metadata": {},
"source": [
"The data for this COVID-19 dataset is stored in a public accessible S3 bucket. You can use the following command to explore the dataset.\n",
"\n",
"!aws s3 ls s3://covid19-lake/ --no-sign-request"
]
},
{
"cell_type": "markdown",
"id": "87f05601-529a-42d4-8cab-ba0b9445e695",
"metadata": {
"tags": []
},
"source": [
"### Read parameters from Cloud Formation stack\n",
"Some of the resources needed for this notebook such as the LLM model endpoint, the AWS Glue database and Glue crawler are created through a cloud formation template. The next block of code extracts the outputs and parameters of the cloud formation stack created from that template to get the value of these parameters.\n",
"\n",
"*The stack name here should match the stack name you used when creating the cloud formation stack.*"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "406e3ede-99aa-46e0-b3aa-eb8b65f6cff5",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# if used a different name while creating the cloud formation stack then change this to match the name you used\n",
"CFN_STACK_NAME = \"cfn-genai-mda\""
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "86a1efa5-a1ae-4384-b149-4103b70fab56",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"stacks = boto3.client('cloudformation').list_stacks()\n",
"stack_found = CFN_STACK_NAME in [stack['StackName'] for stack in stacks['StackSummaries']]"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "c0bda7a4-f2a9-4379-b4a9-8e3e17240d0f",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from typing import List\n",
"def get_cfn_outputs(stackname: str) -> List:\n",
" cfn = boto3.client('cloudformation')\n",
" outputs = {}\n",
" for output in cfn.describe_stacks(StackName=stackname)['Stacks'][0]['Outputs']:\n",
" outputs[output['OutputKey']] = output['OutputValue']\n",
" return outputs\n",
"\n",
"def get_cfn_parameters(stackname: str) -> List:\n",
" cfn = boto3.client('cloudformation')\n",
" params = {}\n",
" for param in cfn.describe_stacks(StackName=stackname)['Stacks'][0]['Parameters']:\n",
" params[param['ParameterKey']] = param['ParameterValue']\n",
" return params\n",
"\n",
"if stack_found is True:\n",
" outputs = get_cfn_outputs(CFN_STACK_NAME)\n",
" params = get_cfn_parameters(CFN_STACK_NAME)\n",
" LLMEndpointName = outputs['LLMEndpointName']\n",
" glue_crawler_name = params['CFNCrawlerName']\n",
" glue_database_name = params['CFNDatabaseName']\n",
" glue_databucket_name = params['DataBucketName']\n",
" region = outputs['Region']\n",
" print(f\"cfn outputs={outputs}\\nparams={params}\")\n",
"else:\n",
" print(\"Recheck our cloudformation stack name\")"
]
},
{
"cell_type": "markdown",
"id": "59c5c0b3-d32e-41ff-85fe-dc80586fa737",
"metadata": {
"tags": []
},
"source": [
"### Copy the sample dataset to your S3 bucket"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "ceedd61d-9c21-45b4-b35e-69e5cd047f11",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!aws s3 cp --recursive s3://covid19-lake/rearc-covid-19-testing-data/json/states_daily/ s3://{glue_databucket_name}/covid-dataset/"
]
},
{
"cell_type": "markdown",
"id": "7dbd2b73-cec1-4dbf-8a94-692eb04e7979",
"metadata": {
"tags": []
},
"source": [
"### Run the crawler"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "5ccbd062-0918-4dcf-a4c2-45bfa81e56c7",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile python_glueworkshop.py\n",
"import boto3\n",
"import argparse\n",
"import time\n",
"\n",
"argParser = argparse.ArgumentParser()\n",
"argParser.add_argument(\"-c\", \"--glue_crawler_name\", help=\"script help\")\n",
"args = argParser.parse_args()\n",
"print(args.glue_crawler_name )\n",
"client = boto3.client('glue')\n",
"crawler_name=args.glue_crawler_name\n",
"\n",
"def get_crawler_status(crawler_name):\n",
" # Create a Glue client\n",
" glue_client = boto3.client('glue')\n",
"\n",
" # Get the crawler details\n",
" response = glue_client.get_crawler(Name=crawler_name)\n",
"\n",
" # Extract the crawler state\n",
" crawler_state = response['Crawler']['State']\n",
"\n",
" return crawler_state\n",
"\n",
"# This is the command to start the Crawler\n",
"try:\n",
" response = client.start_crawler(Name=crawler_name )\n",
" print(\"Successfully started crawler. The crawler may take 2-5 mins to detect the schema.\")\n",
"\n",
" while True:\n",
" # Get the crawler status\n",
" status = get_crawler_status(crawler_name)\n",
"\n",
" # Print the crawler status\n",
" print(f\"Crawler '{crawler_name}' status: {status}\")\n",
"\n",
" if status == 'READY': # Replace 'READY' with the desired completed state\n",
" break # Exit the loop if the desired state is reached\n",
"\n",
" time.sleep(10) # Sleep for 10 seconds before checking the status again\n",
" \n",
"except:\n",
" print(\"error in starting crawler. Check the logs for the error details.\")\n"
]
},
{
"cell_type": "markdown",
"id": "d1392725-610f-43c1-8fd9-b5f52c4585b1",
"metadata": {},
"source": [
"Execute the python script by passing the glue crawler name from the cloudformation stack output."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "75d0fa47-1651-4cff-802a-22ae6438a09c",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!python python_glueworkshop.py -c {glue_crawler_name}"
]
},
{
"cell_type": "markdown",
"id": "4132ffc3-6947-49b6-b627-fae3df870b88",
"metadata": {
"tags": []
},
"source": [
"Before proceeding to the next step, check the status of the crawler. It should change from RUNNING to READY. "
]
},
{
"cell_type": "markdown",
"id": "b51d1d0e-33fb-46ca-b82f-6294ea867cae",
"metadata": {
"tags": []
},
"source": [
"### Step 1 - Connect to databases using SQL Alchemy. \n",
"\n",
"Under the hood, LangChain uses SQLAlchemy to connect to SQL databases. The SQLDatabaseChain can therefore be used with any SQL dialect supported by SQLAlchemy, \n",
"such as MS SQL, MySQL, MariaDB, PostgreSQL, Oracle SQL, and SQLite. Please refer to the SQLAlchemy documentation for more information about requirements for connecting to your database. \n"
]
},
{
"cell_type": "markdown",
"id": "e5f5ce28-9b33-4061-8655-2b297d5c24a2",
"metadata": {
"tags": []
},
"source": [
"**Important**: The code below establishes a database connection for data sources and Large Language Models. Please note that the solution will only work if the database connection for your sources is defined in the cell below. Please refer to the Pre-requisites section. If your use case requires data from Aurora MySQL alone, then please comment out other data sources. Furthermore, please update the cluster details and variables for Aurora MySQL accordingly."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "1583cade",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"#define connections\n",
"\n",
"#LLM \n",
"llm=SagemakerEndpoint(\n",
" endpoint_name=LLMEndpointName, \n",
" region_name=region,\n",
" model_kwargs={\"temperature\":1e-10},\n",
" content_handler=content_handler\n",
" )\n",
"\n",
"#S3\n",
"# connect to s3 using athena\n",
"## athena variables\n",
"connathena=f\"athena.{region}.amazonaws.com\" \n",
"portathena='443' #Update, if port is different\n",
"schemaathena=glue_database_name #from cfn params\n",
"s3stagingathena=f's3://{glue_databucket_name}/athenaresults/'#from cfn params\n",
"wkgrpathena='primary'#Update, if workgroup is different\n",
"# tablesathena=['dataset']#[
\n"
]
},
{
"cell_type": "markdown",
"id": "5d0297e0-f2dd-464b-9254-6693c45ebafc",
"metadata": {
"tags": []
},
"source": [
"Step 1. Connection to various channels through which LLMs can talk to your data. These channels include:\n",
"\n",
" - RedShift Serverless - to connect to datastore 'tickit'(ticket is referred as tickit in the sample data store) to retrieve information regarding ticket sales.\n",
" - Aurora - MySQL Serverless - to connect to datastore that hosts information about the employees.\n",
" - S3/Athena - to connect to the SageMaker's offline feature store on claims information. \n",
" - Snowflake - to connect to stocks related data residing in finance schema of 3rd party software.\n",
" - APIs - to connect to meteo(in this example we use Langchain's sample dataset on meteo) to retrieve weather information.\n",
" \n",
"Step 2. Usage of Dynamic generation of prompt templates by populating metadata of the tables using Glue Data Catalog(GDC) as context. GDC was populated by running a crawler on the databases. Refer to the information here to create and run a glue crawler. In case of api, a line item was created in GDC data extract.\n",
"\n",
"Step 3. Define Functions to 1/ determine the best data channel to answer the user query, 2/ Generate response to user query\n",
"\n",
"Step 4. Apply user query to LLM and Langchain to determine the data channel. After determining the data channel, run the Langchain SQL Database chain to convert 'text to sql' and run the query against the source data channel. \n",
"\n",
"Finally, display the results.\n"
]
},
{
"cell_type": "markdown",
"id": "e0986ea2-f794-431f-a341-b94f0118cb7d",
"metadata": {
"tags": []
},
"source": [
"### Pre-requisites:\n",
"1. Use kernel Base Python 3.0.\n",
"2. Deploy resources using the cloudformation template mda-llm-cfn.yml.\n",
"\n",
"[OPTIONAL] - If need is to add any of the sources below, then uncomment code in the relevant sections.\n",
"\n",
"1. Setup [Aurora MySQL Serverless database](https://aws.amazon.com/getting-started/hands-on/building-serverless-applications-with-amazon-aurora-serverless/?ref=gsrchandson). Load sample dataset for Human Resource department. Use this notebook to load the data into Aurora MySQL.\n",
"2. Setup [Redshift Serverless](https://catalog.workshops.aws/redshift-immersion/en-US/lab1). Load sample data for Sales & Marketing. For example, 'sample data dev' for 'tickit' dataset available in RedShift examples.\n",
"3. Setup External database. In this case, we are using Snowflake account and populating stocks data. Use this notebook to load the data into Snowflake.\n",
"4. Add/modify the [Glue Crawler](https://catalog.us-east-1.prod.workshops.aws/workshops/71b5bdcf-7eb1-4549-b851-66adc860cd04/en-US/2-studio/1-crawler) on all the databases mentioned above. \n",
"\n",
"**Note - This notebook was tested on kernel - conda_python3 in Region us-east-1**"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "9556eddc-8e45-4e42-9157-213316ec468a",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile requirements.txt\n",
"sqlalchemy==1.4.47\n",
"snowflake-sqlalchemy\n",
"langchain==0.0.166\n",
"sqlalchemy-aurora-data-api\n",
"PyAthena[SQLAlchemy]==2.25.2\n",
"redshift-connector==2.0.910\n",
"sqlalchemy-redshift==0.8.14"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "b55d516c",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"!pip install -r requirements.txt"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "91c153cd",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"import json\n",
"import boto3\n",
"\n",
"import sqlalchemy\n",
"from sqlalchemy import create_engine\n",
"from snowflake.sqlalchemy import URL\n",
"\n",
"from langchain.docstore.document import Document\n",
"from langchain import PromptTemplate,SagemakerEndpoint,SQLDatabase, SQLDatabaseChain, LLMChain\n",
"from langchain.llms.sagemaker_endpoint import LLMContentHandler\n",
"from langchain.chains.question_answering import load_qa_chain\n",
"from langchain.prompts.prompt import PromptTemplate\n",
"from langchain.chains import SQLDatabaseSequentialChain\n",
"\n",
"from langchain.chains.api.prompt import API_RESPONSE_PROMPT\n",
"from langchain.chains import APIChain\n",
"from langchain.prompts.prompt import PromptTemplate\n",
"\n",
"from langchain.chains.api import open_meteo_docs\n",
"\n",
"from typing import Dict"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "f9c445a3-0f6b-4164-83f4-bd5ddf8d084c",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"#define content handler class for flant5xl model\n",
"from langchain.llms.sagemaker_endpoint import LLMContentHandler\n",
"class ContentHandler(LLMContentHandler):\n",
" content_type = \"application/json\"\n",
" accepts = \"application/json\"\n",
" \n",
" def transform_input(self, prompt, model_kwargs) :\n",
" test = {\"text_inputs\": prompt}\n",
" encoded_json = json.dumps(test).encode(\"utf-8\")\n",
" return encoded_json\n",
" \n",
" def transform_output(self, output):\n",
" response_json = json.loads(output.read().decode(\"utf-8\")).get('generated_texts')\n",
" print(\"response\" , response_json)\n",
" return \"\".join(response_json)\n",
"\n",
"content_handler = ContentHandler()"
]
},
{
"cell_type": "markdown",
"id": "074a4144-4053-46c1-ba39-8f64f8fb9e00",
"metadata": {},
"source": [
"The data for this COVID-19 dataset is stored in a public accessible S3 bucket. You can use the following command to explore the dataset.\n",
"\n",
"!aws s3 ls s3://covid19-lake/ --no-sign-request"
]
},
{
"cell_type": "markdown",
"id": "87f05601-529a-42d4-8cab-ba0b9445e695",
"metadata": {
"tags": []
},
"source": [
"### Read parameters from Cloud Formation stack\n",
"Some of the resources needed for this notebook such as the LLM model endpoint, the AWS Glue database and Glue crawler are created through a cloud formation template. The next block of code extracts the outputs and parameters of the cloud formation stack created from that template to get the value of these parameters.\n",
"\n",
"*The stack name here should match the stack name you used when creating the cloud formation stack.*"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "406e3ede-99aa-46e0-b3aa-eb8b65f6cff5",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# if used a different name while creating the cloud formation stack then change this to match the name you used\n",
"CFN_STACK_NAME = \"cfn-genai-mda\""
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "86a1efa5-a1ae-4384-b149-4103b70fab56",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"stacks = boto3.client('cloudformation').list_stacks()\n",
"stack_found = CFN_STACK_NAME in [stack['StackName'] for stack in stacks['StackSummaries']]"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "c0bda7a4-f2a9-4379-b4a9-8e3e17240d0f",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from typing import List\n",
"def get_cfn_outputs(stackname: str) -> List:\n",
" cfn = boto3.client('cloudformation')\n",
" outputs = {}\n",
" for output in cfn.describe_stacks(StackName=stackname)['Stacks'][0]['Outputs']:\n",
" outputs[output['OutputKey']] = output['OutputValue']\n",
" return outputs\n",
"\n",
"def get_cfn_parameters(stackname: str) -> List:\n",
" cfn = boto3.client('cloudformation')\n",
" params = {}\n",
" for param in cfn.describe_stacks(StackName=stackname)['Stacks'][0]['Parameters']:\n",
" params[param['ParameterKey']] = param['ParameterValue']\n",
" return params\n",
"\n",
"if stack_found is True:\n",
" outputs = get_cfn_outputs(CFN_STACK_NAME)\n",
" params = get_cfn_parameters(CFN_STACK_NAME)\n",
" LLMEndpointName = outputs['LLMEndpointName']\n",
" glue_crawler_name = params['CFNCrawlerName']\n",
" glue_database_name = params['CFNDatabaseName']\n",
" glue_databucket_name = params['DataBucketName']\n",
" region = outputs['Region']\n",
" print(f\"cfn outputs={outputs}\\nparams={params}\")\n",
"else:\n",
" print(\"Recheck our cloudformation stack name\")"
]
},
{
"cell_type": "markdown",
"id": "59c5c0b3-d32e-41ff-85fe-dc80586fa737",
"metadata": {
"tags": []
},
"source": [
"### Copy the sample dataset to your S3 bucket"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "ceedd61d-9c21-45b4-b35e-69e5cd047f11",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!aws s3 cp --recursive s3://covid19-lake/rearc-covid-19-testing-data/json/states_daily/ s3://{glue_databucket_name}/covid-dataset/"
]
},
{
"cell_type": "markdown",
"id": "7dbd2b73-cec1-4dbf-8a94-692eb04e7979",
"metadata": {
"tags": []
},
"source": [
"### Run the crawler"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "5ccbd062-0918-4dcf-a4c2-45bfa81e56c7",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile python_glueworkshop.py\n",
"import boto3\n",
"import argparse\n",
"import time\n",
"\n",
"argParser = argparse.ArgumentParser()\n",
"argParser.add_argument(\"-c\", \"--glue_crawler_name\", help=\"script help\")\n",
"args = argParser.parse_args()\n",
"print(args.glue_crawler_name )\n",
"client = boto3.client('glue')\n",
"crawler_name=args.glue_crawler_name\n",
"\n",
"def get_crawler_status(crawler_name):\n",
" # Create a Glue client\n",
" glue_client = boto3.client('glue')\n",
"\n",
" # Get the crawler details\n",
" response = glue_client.get_crawler(Name=crawler_name)\n",
"\n",
" # Extract the crawler state\n",
" crawler_state = response['Crawler']['State']\n",
"\n",
" return crawler_state\n",
"\n",
"# This is the command to start the Crawler\n",
"try:\n",
" response = client.start_crawler(Name=crawler_name )\n",
" print(\"Successfully started crawler. The crawler may take 2-5 mins to detect the schema.\")\n",
"\n",
" while True:\n",
" # Get the crawler status\n",
" status = get_crawler_status(crawler_name)\n",
"\n",
" # Print the crawler status\n",
" print(f\"Crawler '{crawler_name}' status: {status}\")\n",
"\n",
" if status == 'READY': # Replace 'READY' with the desired completed state\n",
" break # Exit the loop if the desired state is reached\n",
"\n",
" time.sleep(10) # Sleep for 10 seconds before checking the status again\n",
" \n",
"except:\n",
" print(\"error in starting crawler. Check the logs for the error details.\")\n"
]
},
{
"cell_type": "markdown",
"id": "d1392725-610f-43c1-8fd9-b5f52c4585b1",
"metadata": {},
"source": [
"Execute the python script by passing the glue crawler name from the cloudformation stack output."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "75d0fa47-1651-4cff-802a-22ae6438a09c",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!python python_glueworkshop.py -c {glue_crawler_name}"
]
},
{
"cell_type": "markdown",
"id": "4132ffc3-6947-49b6-b627-fae3df870b88",
"metadata": {
"tags": []
},
"source": [
"Before proceeding to the next step, check the status of the crawler. It should change from RUNNING to READY. "
]
},
{
"cell_type": "markdown",
"id": "b51d1d0e-33fb-46ca-b82f-6294ea867cae",
"metadata": {
"tags": []
},
"source": [
"### Step 1 - Connect to databases using SQL Alchemy. \n",
"\n",
"Under the hood, LangChain uses SQLAlchemy to connect to SQL databases. The SQLDatabaseChain can therefore be used with any SQL dialect supported by SQLAlchemy, \n",
"such as MS SQL, MySQL, MariaDB, PostgreSQL, Oracle SQL, and SQLite. Please refer to the SQLAlchemy documentation for more information about requirements for connecting to your database. \n"
]
},
{
"cell_type": "markdown",
"id": "e5f5ce28-9b33-4061-8655-2b297d5c24a2",
"metadata": {
"tags": []
},
"source": [
"**Important**: The code below establishes a database connection for data sources and Large Language Models. Please note that the solution will only work if the database connection for your sources is defined in the cell below. Please refer to the Pre-requisites section. If your use case requires data from Aurora MySQL alone, then please comment out other data sources. Furthermore, please update the cluster details and variables for Aurora MySQL accordingly."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "1583cade",
"metadata": {
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"#define connections\n",
"\n",
"#LLM \n",
"llm=SagemakerEndpoint(\n",
" endpoint_name=LLMEndpointName, \n",
" region_name=region,\n",
" model_kwargs={\"temperature\":1e-10},\n",
" content_handler=content_handler\n",
" )\n",
"\n",
"#S3\n",
"# connect to s3 using athena\n",
"## athena variables\n",
"connathena=f\"athena.{region}.amazonaws.com\" \n",
"portathena='443' #Update, if port is different\n",
"schemaathena=glue_database_name #from cfn params\n",
"s3stagingathena=f's3://{glue_databucket_name}/athenaresults/'#from cfn params\n",
"wkgrpathena='primary'#Update, if workgroup is different\n",
"# tablesathena=['dataset']#[