{
"cells": [
{
"cell_type": "markdown",
"id": "796f40c8-b024-4b00-8dce-a633e022c503",
"metadata": {},
"source": [
" ⚠️ PRE-REQUISITE: Please select `Data Science 3.0` and `Python 3` kernel image, instance type `ml.t3.medium` for SageMaker Studio notebook.
"
]
},
{
"cell_type": "markdown",
"id": "71586519-e396-4086-a1bd-b3e9ae2f492c",
"metadata": {},
"source": [
"## Introduction"
]
},
{
"cell_type": "markdown",
"id": "24b51afd-37f8-4bf2-ba74-739e1aff8549",
"metadata": {},
"source": [
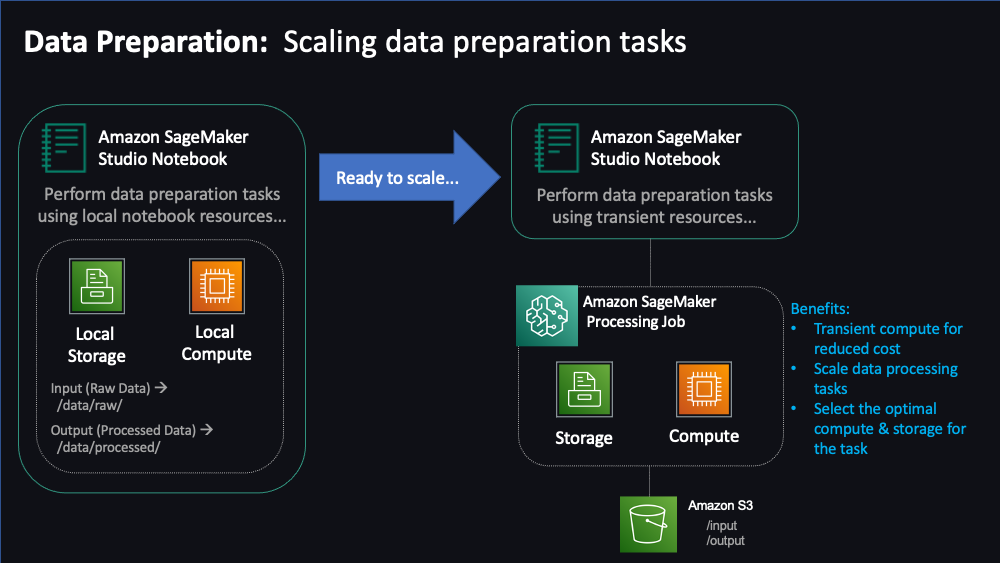
"The first stage in the ML workflow is data preparation. In this notebook, we will explore data preparation on Sagemaker using Ray. Ray is an open-source distributed computing framework designed to accelerate and simplify the development of scalable and efficient machine learning applications. It provides a simple and flexible API for defining and executing [tasks](https://docs.ray.io/en/latest/ray-core/tasks.html) and [actors](https://docs.ray.io/en/latest/ray-core/actors.html) on a cluster of machines, allowing you to easily scale your machine learning workloads from a single machine to thousands of nodes.\n",
"\n",
"Here, we will put on the hat of `Data Scientist`/`Data Engineer` and will perform the tasks of gathering datasets, load them in to a feature store, pre-processing those features to align with our upcoming Training needs. As part of this exercise, we will learn how to bring scale these steps using managed SageMaker processing capabilities. In the last step, we will split the dataset into training, validation and testing sets to be used with out ML workflow.\n",
"\n",
""
]
},
{
"cell_type": "markdown",
"id": "5db66d7f-b9c6-4844-bcc2-e148a9562cd4",
"metadata": {},
"source": [
"To enable you to run these notebooks within a reasonable time (typically less than an hour), the use case is a straightforward regression task: predicting house prices based on a synthetic housing dataset. This dataset contains 8 housing features. Features include year built, number of bedrooms, size of lot, etc...\n",
"\n",
"To begin, we'll import some necessary packages and set up directories for local training and test data. We'll also set up a SageMaker Session to perform various operations, and specify an Amazon S3 bucket to hold input data and output. The default bucket used here is created by SageMaker if it doesn't already exist, and named in accordance with the AWS account ID and AWS Region.\n",
"\n",
"Let's get started!"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "6be4ba9d-c50f-426d-9fbd-3809589f3729",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!pip install -U sagemaker ray modin[ray] pydantic==1.10.10"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "c8c79819-3206-465c-8110-c0904b04cfe2",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"import sagemaker\n",
"import boto3\n",
"import pandas as pd\n",
"from time import strftime\n",
"from sagemaker.sklearn.processing import SKLearnProcessor\n",
"from sagemaker.processing import ProcessingInput, ProcessingOutput\n",
"from sklearn.model_selection import train_test_split\n",
"from sklearn.preprocessing import StandardScaler\n",
"import os\n",
"import time\n",
"from sagemaker.feature_store.feature_group import FeatureGroup\n",
"\n",
"import ray\n",
"from ray.data import Dataset\n",
"from ray.data.preprocessors import StandardScaler\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "4e1f9d3c-961c-4026-8af5-8a1752fb7590",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"local_data_path_ray = \"data/raw/ray/house_pricing.csv\"\n",
"# setting train, validation and test sizes as strings as required by sagemaker arguments\n",
"train_size = 0.6\n",
"val_size = 0.2\n",
"test_size = 0.2\n",
"random_seed = 42 # setting random seed to ensure compatible results over multiple executions\n",
"\n",
"# Useful SageMaker variables\n",
"sess = sagemaker.Session()\n",
"bucket = sess.default_bucket()\n",
"role_arn= sagemaker.get_execution_role()\n",
"region = sess.boto_region_name\n",
"\n",
"# Local data paths\n",
"pipeline_scripts_dir = os.path.join(os.getcwd(), 'pipeline_scripts')\n",
"os.makedirs(pipeline_scripts_dir, exist_ok=True)\n",
"\n",
"processed_dir = os.path.join(os.getcwd(), 'data/processed')\n",
"os.makedirs(processed_dir, exist_ok=True)\n",
"\n",
"processed_train_dir = os.path.join(os.getcwd(), f'{processed_dir}/train')\n",
"os.makedirs(processed_train_dir, exist_ok=True)\n",
"\n",
"processed_validation_dir = os.path.join(os.getcwd(), f'{processed_dir}/validation')\n",
"os.makedirs(processed_validation_dir, exist_ok=True)\n",
"\n",
"processed_test_dir = os.path.join(os.getcwd(), f'{processed_dir}/test')\n",
"os.makedirs(processed_test_dir, exist_ok=True)\n",
"\n",
"# Data paths in S3\n",
"s3_prefix = 'aws-sm-ray-workshop'\n",
"\n",
"# SageMaker Processing variables\n",
"processing_instance_type = 'ml.m5.2xlarge'\n",
"processing_instance_count = 1\n",
"output_path = f's3://{bucket}/{s3_prefix}/data/sm_processed'"
]
},
{
"cell_type": "markdown",
"id": "956a363f-3c59-4492-83eb-ab6820fcb28c",
"metadata": {},
"source": [
"## Starting Ray on a single machine"
]
},
{
"cell_type": "markdown",
"id": "6ae0b1f5-7494-4f5b-af58-3269a8b4d18e",
"metadata": {},
"source": [
"`ray.init()` will attempt to find a Ray instance to connect to automatically. It follows these steps in order:\n",
"\n",
"1. It checks for the `RAY_ADDRESS` OS environment variable.\n",
"2. It looks for the specific address passed to `ray.init(address=)`.\n",
"3. If no address is provided, it connects to the most recent Ray instance that was launched on the same machine using `ray start`.\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "dcf1b786-94e9-4ac3-93c5-fb50afdf3920",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"ray.init(include_dashboard=False)"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "29bc3b12-1a95-46ee-8b7c-7a359f6df125",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# test reading data using ray.data\n",
"dataset = ray.data.read_csv(local_data_path_ray)\n",
"dataset"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "92cc8007-8568-4463-9c5b-e18107a65882",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# Upload raw data to S3\n",
"raw_data_s3_prefix = '{}/data/raw'.format(s3_prefix)\n",
"raw_s3 = sess.upload_data(path=local_data_path_ray, key_prefix=raw_data_s3_prefix)"
]
},
{
"cell_type": "markdown",
"id": "1dc1ce98-4463-4d67-ad95-937bf0a2ba87",
"metadata": {},
"source": [
"We're now ready to run the Processing job for spliting and scaling the data."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "6916c958-e1e8-4041-8db7-6b3d020e5720",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"raw_s3"
]
},
{
"cell_type": "markdown",
"id": "422e84ef-6ef8-466f-97e9-9b428e7e2e47",
"metadata": {
"tags": []
},
"source": [
"## SageMaker Feature Store\n",
" \n",
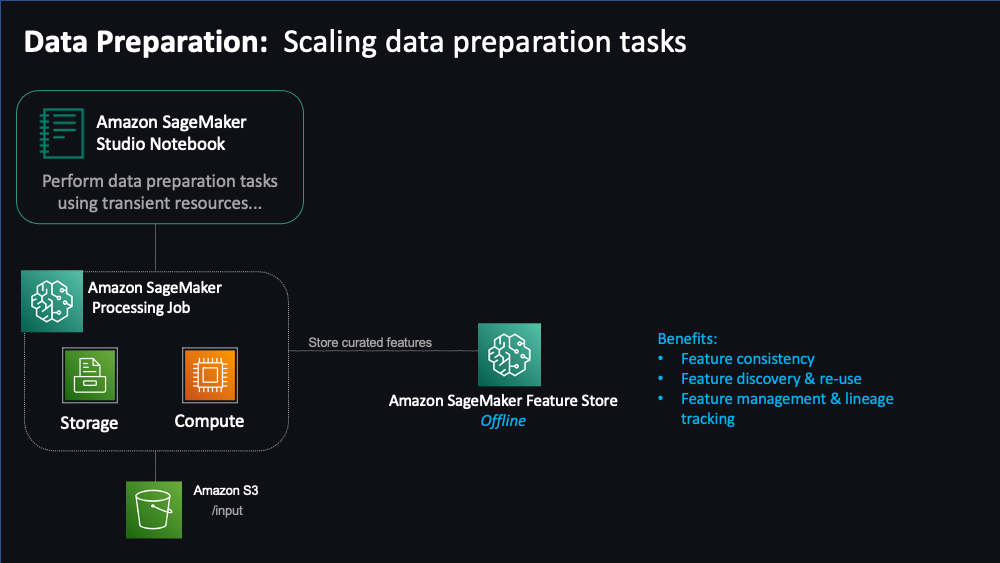
"Features are inputs to ML models used during training and inference. Features are used repeatedly by multiple teams and feature quality is critical to ensure a highly accurate model. Also, when features used to train models offline in batch are made available for real-time inference, it’s hard to keep the two feature stores synchronized. [SageMaker Feature Store](https://docs.aws.amazon.com/sagemaker/latest/dg/feature-store.html) provides a secured and unified store for feature use across the ML lifecycle. \n",
"\n",
"Let's now exchange the storage of our processed data from S3 to SageMaker Feature Store.\n",
"\n",
"\n",
"\n",
"**Feature Groups**\n",
"\n",
"First let's define some feature groups for train, validation and test datasets and a S3 bucket prefix as offline feature store to store your features"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "9f5490ae-200d-4d64-8b84-fc8bd9410694",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# Optional step\n",
"# Delete all Feature Groups that start with the prefix fs-ray-. \n",
"# This is to prevent duplication of feature stores when running this workshop multiple time.\n",
"\n",
"sm_client = boto3.client('sagemaker', region_name=region)\n",
"sagemaker_session = sagemaker.Session(boto3.Session(region_name=region))\n",
"response = sm_client.list_feature_groups(\n",
" NameContains='fs-ray-'\n",
")\n",
"\n",
"for feature in response[\"FeatureGroupSummaries\"]:\n",
" print(f'deleting {feature[\"FeatureGroupName\"]}')\n",
" resp = sm_client.delete_feature_group(\n",
" FeatureGroupName=feature[\"FeatureGroupName\"]\n",
" )"
]
},
{
"cell_type": "markdown",
"id": "a0502598-358f-4e87-bb1a-fcfa3fc5708c",
"metadata": {
"tags": []
},
"source": [
"## SageMaker Processing\n",
" \n",
"To process large amounts of data, we fortunately will not need to write distributed code oursleves. Instead, we can use [SageMaker Processing](https://docs.aws.amazon.com/sagemaker/latest/dg/processing-job.html) which will do all the processing _outside_ of this notebook's resources and will apply our processing script to multiple data files in parallel on SageMaker Processing instances.\n",
" \n",
"Keep in mind that in a typical SageMaker workflow, notebooks are only used for initial model development activities and can be run on relatively inexpensive and less powerful instances. However, to run similar tasks at scale, data scientists require access to more powerful SageMaker managed compute instances for data preparation, training, and model hosting tasks. \n",
"\n",
"SageMaker Processing includes off-the-shelf support for [scikit-learn](https://docs.aws.amazon.com/sagemaker/latest/dg/use-scikit-learn-processing-container.html), [PySpark](https://docs.aws.amazon.com/sagemaker/latest/dg/use-spark-processing-container.html), and [other frameworks](https://docs.aws.amazon.com/sagemaker/latest/dg/processing-job-frameworks.html) like Hugging Face, MXNet, PyTorch, TensorFlow, and XGBoost. You can even a Bring Your Own Container if one our our built-in containers does not suit your use case."
]
},
{
"cell_type": "markdown",
"id": "f353351e-3197-40ae-ba85-9205a738e4d5",
"metadata": {
"tags": []
},
"source": [
"## Processing data using Modin[ray]"
]
},
{
"cell_type": "markdown",
"id": "ce348f92-f47c-431a-bf77-1836b79f04e8",
"metadata": {},
"source": [
"`Modin[Ray]`is an open-source Python library that integrates with Ray to provide a scalable and efficient way to process large datasets. It leverages parallel and distributed computing to accelerate data analysis tasks, making it easier to work with big data.\n",
"No prior knowledge of hardware resources or data distribution is required when using Modin. It is a drop-in replacement for pandas and offers a significant performance boost without any modifications to existing pandas notebooks, even on a single machine. Simply update the import statement and leverage Modin's capabilities, similar to how you would with pandas."
]
},
{
"cell_type": "markdown",
"id": "ab7d751a-1d81-4ac4-8524-68a457e1868d",
"metadata": {
"tags": []
},
"source": [
"Note that in the Featurestore class is decorated with @ray.remote. This makes it a `ray actor`. An actor in Ray is similar to an object in object-oriented programming but with some important differences. Each actor has its own state, which can include variables, data structures, and methods. Actors communicate with each other by sending messages asynchronously, allowing for concurrent and parallel execution. This messaging mechanism is built on top of Ray's underlying task execution engine, enabling efficient and scalable distributed computing."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "7e75769f-2bc7-49b9-9097-c17cc3cc3622",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile ./pipeline_scripts/feature-store/script-fs.py\n",
"\n",
"import subprocess\n",
"import sys\n",
"subprocess.check_call([sys.executable, '-m', 'pip', 'install', 'sagemaker', 'ray', 'modin[ray]', 'pydantic==1.10.10'])\n",
"\n",
"from sagemaker.feature_store.feature_group import FeatureGroup\n",
"import time\n",
"import argparse\n",
"import os\n",
"import numpy as np\n",
"from sklearn.preprocessing import StandardScaler\n",
"from sklearn.model_selection import train_test_split\n",
"import time\n",
"import datetime\n",
"import sagemaker\n",
"import boto3\n",
"import glob\n",
"import modin.pandas as pd\n",
"import ray\n",
"\n",
"########### BEGIN ACTOR #############\n",
"\n",
"@ray.remote(num_cpus=0.5)\n",
"class Featurestore:\n",
" \n",
" def __init__(self):\n",
" pass\n",
" \n",
" def ingest_features(self,feature_group_name, df, region):\n",
" \"\"\"\n",
" Ingest features to Feature Store Group\n",
" Args:\n",
" feature_group_name (str): Feature Group Name\n",
" data_path (str): Path to the train/validation/test data in CSV format.\n",
" \"\"\"\n",
" featurestore_runtime_client = boto3.client('sagemaker-featurestore-runtime', region_name=region)\n",
" for index, row in df.iterrows(): \n",
" try:\n",
" featurestore_runtime_client.put_record(\n",
" FeatureGroupName=feature_group_name,\n",
" Record=[\n",
" {\n",
" 'FeatureName': 'record_id',\n",
" 'ValueAsString': str(int(row['record_id']))\n",
" },\n",
" {\n",
" 'FeatureName': 'event_time',\n",
" 'ValueAsString': str(row['event_time'])\n",
" },\n",
" {\n",
" 'FeatureName': 'NUM_BATHROOMS',\n",
" 'ValueAsString': str(row['NUM_BATHROOMS'])\n",
" },\n",
" {\n",
" 'FeatureName': 'NUM_BEDROOMS',\n",
" 'ValueAsString': str(row['NUM_BEDROOMS'])\n",
" },\n",
" {\n",
" 'FeatureName': 'FRONT_PORCH',\n",
" 'ValueAsString': str(row['FRONT_PORCH'])\n",
" },\n",
" {\n",
" 'FeatureName': 'LOT_ACRES',\n",
" 'ValueAsString': str(row['LOT_ACRES'])\n",
" },\n",
" {\n",
" 'FeatureName': 'DECK',\n",
" 'ValueAsString': str(row['DECK'])\n",
" },\n",
" {\n",
" 'FeatureName': 'SQUARE_FEET',\n",
" 'ValueAsString': str(row['SQUARE_FEET'])\n",
" },\n",
" {\n",
" 'FeatureName': 'YEAR_BUILT',\n",
" 'ValueAsString': str(row['YEAR_BUILT'])\n",
" },\n",
" {\n",
" 'FeatureName': 'GARAGE_SPACES',\n",
" 'ValueAsString': str(row['GARAGE_SPACES'])\n",
" },\n",
" {\n",
" 'FeatureName': 'PRICE',\n",
" 'ValueAsString': str(int(row['PRICE']))\n",
" },\n",
" ],\n",
" TargetStores=[\n",
" 'OfflineStore'\n",
" ]\n",
" )\n",
" except Exception as e:\n",
" print(f\"An error occurred: {e}\\nFailed to process record number {index} for feature group {feature_group_name}\");\n",
"\n",
" return\n",
" \n",
"########### END ACTOR #############\n",
"\n",
"\n",
"def wait_for_feature_group_creation_complete(feature_group):\n",
" \"\"\"\n",
" Function that waits for feature group to be created in SageMaker Feature Store\n",
" Args:\n",
" feature_group (sagemaker.feature_store.feature_group.FeatureGroup): Feature Group\n",
" \"\"\"\n",
" status = feature_group.describe().get('FeatureGroupStatus')\n",
" print(f'Initial status: {status}')\n",
" while status == 'Creating':\n",
" print(f'Waiting for feature group: {feature_group.name} to be created ...')\n",
" time.sleep(5)\n",
" status = feature_group.describe().get('FeatureGroupStatus')\n",
" if status != 'Created':\n",
" raise SystemExit(f'Failed to create feature group {feature_group.name}: {status}')\n",
" print(f'FeatureGroup {feature_group.name} was successfully created.')\n",
"\n",
"def create_feature_group(feature_group_name, prefix, role_arn, region):\n",
" \"\"\"\n",
" Create Feature Store Group\n",
" Args:\n",
" feature_group_name (str): Feature Store Group Name\n",
" sagemaker_session (sagemaker.session.Session): sagemaker session\n",
" df (pandas.DataFrame): dataframe to injest used to create features definition\n",
" prefix (str): geature group prefix (train/validation or test)\n",
" role_arn (str): role arn to create feature store\n",
" Returns:\n",
" fs_group (sagemaker.feature_store.feature_group.FeatureGroup): Feature Group\n",
" \"\"\"\n",
" sm_client = boto3.client('sagemaker', region_name=region)\n",
" sagemaker_session = sagemaker.Session(boto3.Session(region_name=region))\n",
"\n",
" default_bucket = sagemaker_session.default_bucket()\n",
"\n",
" # Search to see if the Feature Group already exists\n",
" results = sm_client.search(\n",
" Resource=\"FeatureGroup\",\n",
" SearchExpression={\n",
" 'Filters': [\n",
" {\n",
" 'Name': 'FeatureGroupName',\n",
" 'Operator': 'Equals',\n",
" 'Value': feature_group_name\n",
" },\n",
" ]\n",
" }\n",
" )\n",
"\n",
" # If a FeatureGroup was not found with the name, create one\n",
" if not results['Results']:\n",
" sm_client.create_feature_group(\n",
" FeatureGroupName=feature_group_name,\n",
" RecordIdentifierFeatureName='record_id',\n",
" EventTimeFeatureName='event_time',\n",
" OnlineStoreConfig={\n",
" \"EnableOnlineStore\": False\n",
" },\n",
" OfflineStoreConfig={\n",
" \"S3StorageConfig\": {\n",
" \"S3Uri\": f's3://{default_bucket}/{prefix}', \n",
" }, \n",
" },\n",
" FeatureDefinitions=[\n",
" {\n",
" 'FeatureName': 'record_id',\n",
" 'FeatureType': 'Integral'\n",
" },\n",
" {\n",
" 'FeatureName': 'event_time',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'NUM_BATHROOMS',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'NUM_BEDROOMS',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'FRONT_PORCH',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'LOT_ACRES',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'DECK',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'SQUARE_FEET',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'YEAR_BUILT',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'GARAGE_SPACES',\n",
" 'FeatureType': 'Fractional'\n",
" },\n",
" {\n",
" 'FeatureName': 'PRICE',\n",
" 'FeatureType': 'Integral'\n",
" },\n",
" ],\n",
" RoleArn=role_arn\n",
" )\n",
"\n",
" fs_group = FeatureGroup(\n",
" name=feature_group_name, \n",
" sagemaker_session=sagemaker_session\n",
" )\n",
"\n",
" wait_for_feature_group_creation_complete(fs_group)\n",
" return fs_group\n",
"\n",
"\n",
"def prepare_df_for_feature_store(df):\n",
" \"\"\"\n",
" Add event time and record id to df in order to store it in SageMaker Feature Store\n",
" Args:\n",
" df (pandas.DataFrame): data to be prepared\n",
" Returns:\n",
" df (pandas.DataFrame): dataframe with event time and record id\n",
" \"\"\"\n",
" print(f'Preparing data for Feature Store..')\n",
" current_time_sec = int(round(time.time()))\n",
" # create event time\n",
" df['event_time'] = pd.Series([current_time_sec]*len(df), dtype=\"float64\")\n",
" # create record id from index\n",
" df['record_id'] = df.reset_index().index\n",
" return df\n",
"\n",
"def read_csv(path, num_actors):\n",
" \"\"\"\n",
" Read all the CSV files with in a given directory\n",
" IMPORTANT: All CSVs should have the same schema\n",
" Args:\n",
" path: the path in which the input file exist\n",
" Returns:\n",
" df (pandas.DataFrame): dataframe with CSV data\n",
" \"\"\"\n",
" \n",
" csv_files = glob.glob(os.path.join(path, \"*.csv\"))\n",
" print(f\"found {len(csv_files)} files\")\n",
" frames = []\n",
" # loop over the list of csv files\n",
" for f in csv_files:\n",
" # read the csv file\n",
" df = pd.read_csv(f)\n",
" frames.append(df)\n",
"\n",
" data = pd.concat(frames)\n",
" data = prepare_df_for_feature_store(data)\n",
" # Split into partitions\n",
" partitions = [ray.put(part) for part in np.array_split(data, num_actors)]\n",
" return partitions\n",
" \n",
"\n",
"def read_parameters():\n",
" \"\"\"\n",
" Read job parameters\n",
" Returns:\n",
" (Namespace): read parameters\n",
" \"\"\"\n",
" parser = argparse.ArgumentParser()\n",
" parser.add_argument('--feature_group_name', type=str, default='fs-ray-synthetic-house-price')\n",
" parser.add_argument('--bucket_prefix', type=str, default='aws-ray-mlops-workshop/feature-store')\n",
" parser.add_argument('--num_actors', type=int, default=4)\n",
" parser.add_argument('--region', type=str, default='us-east-1')\n",
" parser.add_argument('--role_arn', type=str)\n",
" params, _ = parser.parse_known_args()\n",
" return params\n",
"\n",
"start = time.time() \n",
"print(f\"===========================================================\")\n",
"print(f\"Starting Feature Store Ingestion\")\n",
"print(f\"Reading parameters\")\n",
"\n",
"ray.init(runtime_env={'env_vars': {'__MODIN_AUTOIMPORT_PANDAS__': '1'}})\n",
"\n",
"# reading job parameters\n",
"args = read_parameters()\n",
"print(f\"Parameters read: {args}\")\n",
"\n",
"\n",
"create_feature_group(\n",
" args.feature_group_name, \n",
" f'{args.bucket_prefix}/synthetic-housing-price-data',\n",
" args.role_arn,\n",
" args.region\n",
" )\n",
"\n",
"\n",
"# set input path\n",
"input_data_path = \"/opt/ml/processing/input/\"\n",
"\n",
"input_partitions = read_csv(input_data_path, args.num_actors)\n",
"\n",
"\n",
"# Start actors and assign partitions in a loop\n",
"actors = [Featurestore.remote() for _ in range(args.num_actors)]\n",
"results = []\n",
"for actor, partition in zip(actors, input_partitions):\n",
" results.append(actor.ingest_features.remote(args.feature_group_name, partition, args.region))\n",
" \n",
"#for actor_state in ray.util.state.list_actors():\n",
"# print(actor_state.node_id, actor_state.pid, actor_state.name, actor_state.state)\n",
"\n",
"ray.get(results)\n",
"\n",
"taken = time.time() - start\n",
"print(f\"Ending Feature Store Ingestion\")\n",
"print(f\"TOTAL TIME TAKEN: {taken:.2f} seconds\")\n",
"print(f\"===========================================================\")"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "29446ab5-779b-4ec1-9c7c-cf2f11b8f9c1",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"sklearn_processor = SKLearnProcessor(\n",
" framework_version='1.0-1',\n",
" role=role_arn,\n",
" instance_type=processing_instance_type,\n",
" instance_count=processing_instance_count\n",
")"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "a66c6150-39af-44f9-bc50-01c307544297",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"time_str = '-' + time.strftime('%Y-%m-%d-%H-%M-%S')\n",
"feature_group_name = \"fs-ray-synthetic_home-price\"+time_str\n",
"bucket_prefix = f'{s3_prefix}/data/feature-store'"
]
},
{
"cell_type": "markdown",
"id": "fce2bb71-42d6-426f-b2df-221baf058879",
"metadata": {},
"source": [
"### Ray parallel processing\n",
"Note that we specify the num of actors as a parameter to the processing job. This determines the number of instances of the actor that will run the data procesing job. Also note the num_cpu parameter specified on top of the actor class. This paramter controls the amount of CPU resources allocated per actor instance. In this case it is 0.25 CPU. This will enable fractional CPU allocation in increments of 0.25 CPUs and help pack more actors per node. The ability to tune fractional CPU resources lets you maximize overall resource usage efficiency in a Ray cluster. The optimal allocation balances parallelism, throughput and schedulability across all tasks."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "e09830fd-95ed-4c80-b563-c0ce66f14b7e",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from IPython.core.display import display, HTML\n",
"\n",
"# code=can be a s3 uri for the input script\n",
"job_name = f\"processing-with-fs-{strftime('%Y-%m-%d-%H-%M-%S')}\"\n",
"\n",
"display(\n",
" HTML(\n",
" 'Review Feature Store Processing Job'.format(\n",
" region, job_name\n",
" )\n",
" )\n",
")\n",
"\n",
"sklearn_processor.run(\n",
" code='pipeline_scripts/feature-store/script-fs.py',\n",
" job_name=job_name,\n",
" inputs=[\n",
" ProcessingInput(\n",
" source=raw_s3,\n",
" destination='/opt/ml/processing/input',\n",
" s3_data_distribution_type='ShardedByS3Key'\n",
" )\n",
" ],\n",
" # notice that all arguments passed to a SageMaker processing job should be strings as they are transformed to command line parameters.\n",
" # Your read_parameters function will handle the data types for your code \n",
" arguments=[\n",
" \"--feature_group_name\", feature_group_name,\n",
" \"--num_actors\", '10',\n",
" \"--bucket_prefix\", bucket_prefix,\n",
" \"--role_arn\", role_arn,\n",
" \"--region\", region\n",
" ]\n",
")\n",
"\n",
"preprocessing_job_description = sklearn_processor.jobs[-1].describe()"
]
},
{
"cell_type": "markdown",
"id": "a329f70a-e703-4849-b0bf-2eff0916a2a2",
"metadata": {
"tags": []
},
"source": [
"### Splitting and Scaling data\n",
"\n",
"Now let's process our data for a Machine Learning model. \n",
"\n",
"We will create a script that will be will split the data into train, validation and test datasets using feature available with in the `ray.data` library. Then scale all columns other than the target column using a standard scaler from `ray.data.preprocessors`. \n",
"\n",
"Ray provides a `ray.data.Dataset` API for working with distributed datasets. This API allows you to perform parallel processing on large datasets by automatically partitioning the data into blocks and executing operations on these blocks in parallel.\n",
"\n",
"When you create a `ray.data.Dataset` object, the data is automatically partitioned into blocks based on the size of the data and the available resources in your Ray cluster. Each block is stored as a separate Ray object, allowing multiple tasks to operate on different blocks in parallel.\n",
"\n",
"When you perform operations on a `ray.data.Dataset`, such as applying a transformation or computing an aggregation, Ray automatically schedules tasks to execute these operations on the individual blocks in parallel. The results of these operations are then combined to produce the final result.\n",
"\n",
"Besides preparing a Ray preprocessing script, we also need to prepare a requirements.txt with ray listed."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "c761e639-4e70-47c5-b8d3-cf8be742fa7f",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile ./pipeline_scripts/preprocessing/script.py\n",
"import subprocess\n",
"import sys\n",
"subprocess.check_call([sys.executable, '-m', 'pip', 'install', 'sagemaker','ray', 'pyarrow >= 6.0.1'])\n",
"\n",
"import argparse\n",
"import os\n",
"\n",
"import sagemaker\n",
"# Experiments\n",
"from sagemaker.session import Session\n",
"from sagemaker.feature_store.feature_group import FeatureGroup\n",
"\n",
"import boto3\n",
"import ray\n",
"from ray.air.config import ScalingConfig\n",
"from ray.data import Dataset\n",
"from ray.data.preprocessors import StandardScaler\n",
"\n",
"def read_parameters():\n",
" \"\"\"\n",
" Read job parameters\n",
" Returns:\n",
" (Namespace): read parameters\n",
" \"\"\"\n",
" parser = argparse.ArgumentParser()\n",
" parser.add_argument('--feature_group_name', type=str, default='fs-synthetic-house-price')\n",
" parser.add_argument('--train_size', type=float, default=0.6)\n",
" parser.add_argument('--val_size', type=float, default=0.2)\n",
" parser.add_argument('--test_size', type=float, default=0.2)\n",
" parser.add_argument('--random_state', type=int, default=42)\n",
" parser.add_argument('--target_col', type=str, default='PRICE')\n",
" parser.add_argument('--region', type=str, default='us-east-1')\n",
" params, _ = parser.parse_known_args()\n",
" return params\n",
"\n",
"def split_dataset(dataset, train_size, val_size, test_size, random_state=None):\n",
" \"\"\"\n",
" Split dataset into train, validation and test samples\n",
" Args:\n",
" dataset (ray.data.Dataset): input data\n",
" train_size (float): ratio of data to use as training dataset\n",
" val_size (float): ratio of data to use as validation dataset\n",
" test_size (float): ratio of data to use as test dataset\n",
" random_state (int): Pass an int for reproducible output across multiple function calls.\n",
" Returns:\n",
" train_set (ray.data.Dataset): train dataset\n",
" val_set (ray.data.Dataset): validation dataset\n",
" test_set (ray.data.Dataset): test dataset\n",
" \"\"\"\n",
" if (train_size + val_size + test_size) != 1.0:\n",
" raise ValueError(\"train_size, val_size and test_size must sum up to 1.0\")\n",
" \n",
" # Shuffle this dataset with a fixed random seed.\n",
" shuffled_ds = dataset.random_shuffle(seed=random_state)\n",
" # Split the data into train, validation and test datasets\n",
" train_set, val_set, test_set = shuffled_ds.split_proportionately([train_size, val_size])\n",
" \n",
" # Sanity check\n",
" # IMPORTANT!!! Do not include this for large datasets as this can be an expensive operation\n",
" train_perc = int((train_set.count()/shuffled_ds.count()) * 100)\n",
" print(f\"Training size: {train_set.count()} - {train_perc}% of total\")\n",
" val_perc = int((val_set.count()/shuffled_ds.count()) * 100)\n",
" print(f\"Val size: {val_set.count()} - {val_perc}% of total\")\n",
" test_perc = int((test_set.count()/shuffled_ds.count()) * 100)\n",
" print(f\"Test size: {test_set.count()} - {test_perc}% of total\")\n",
" return train_set, val_set, test_set\n",
"\n",
"def scale_dataset(train_set, val_set, test_set, target_col):\n",
" \"\"\"\n",
" Fit StandardScaler to train_set and apply it to val_set and test_set\n",
" Args:\n",
" train_set (ray.data.Dataset): train dataset\n",
" val_set (ray.data.Dataset): validation dataset\n",
" test_set (ray.data.Dataset): test dataset\n",
" target_col (str): target col\n",
" Returns:\n",
" train_transformed (ray.data.Dataset): train data scaled\n",
" val_transformed (ray.data.Dataset): val data scaled\n",
" test_transformed (ray.data.Dataset): test data scaled\n",
" \"\"\"\n",
" \n",
" tranform_cols = dataset.columns()\n",
" # Remove the target columns from being scaled\n",
" tranform_cols.remove(target_col)\n",
" # set up a standard scaler\n",
" standard_scaler = StandardScaler(tranform_cols)\n",
" # fit scaler to training dataset\n",
" print(\"Fitting scaling to training data and transforming dataset...\")\n",
" train_set_transformed = standard_scaler.fit_transform(train_set)\n",
" # apply scaler to validation and test datasets\n",
" print(\"Transforming validation and test datasets...\")\n",
" val_set_transformed = standard_scaler.transform(val_set)\n",
" test_set_transformed = standard_scaler.transform(test_set)\n",
" return train_set_transformed, val_set_transformed, test_set_transformed\n",
"\n",
"def load_dataset(feature_group_name, region):\n",
" \"\"\"\n",
" Loads the data as a ray dataset from the offline featurestore S3 location\n",
" Args:\n",
" feature_group_name (str): name of the feature group\n",
" Returns:\n",
" ds (ray.data.dataset): Ray dataset the contains the requested dat from the feature store\n",
" \"\"\"\n",
" session = sagemaker.Session(boto3.Session(region_name=region))\n",
" fs_group = FeatureGroup(\n",
" name=feature_group_name, \n",
" sagemaker_session=session\n",
" )\n",
"\n",
" fs_data_loc = fs_group.describe().get(\"OfflineStoreConfig\").get(\"S3StorageConfig\").get(\"ResolvedOutputS3Uri\")\n",
" \n",
" # Drop columns added by the feature store\n",
" # Since these are not related to the ML problem at hand\n",
" cols_to_drop = [\"record_id\", \"event_time\",\"write_time\", \n",
" \"api_invocation_time\", \"is_deleted\", \n",
" \"year\", \"month\", \"day\", \"hour\"] \n",
"\n",
" ds = ray.data.read_parquet(fs_data_loc)\n",
" ds = ds.drop_columns(cols_to_drop)\n",
" print(f\"{fs_data_loc} count is {ds.count()}\")\n",
" return ds\n",
"\n",
"print(f\"===========================================================\")\n",
"print(f\"Starting pre-processing\")\n",
"print(f\"Reading parameters\")\n",
"\n",
"# reading job parameters\n",
"args = read_parameters()\n",
"print(f\"Parameters read: {args}\")\n",
"\n",
"# set output paths\n",
"train_data_path = \"/opt/ml/processing/output/train\"\n",
"val_data_path = \"/opt/ml/processing/output/validation\"\n",
"test_data_path = \"/opt/ml/processing/output/test\"\n",
"\n",
"try:\n",
" os.makedirs(train_data_path)\n",
" os.makedirs(val_data_path)\n",
" os.makedirs(test_data_path)\n",
"except:\n",
" pass\n",
"\n",
"# read data input\n",
"dataset = load_dataset(args.feature_group_name, args.region)\n",
"\n",
"# split dataset into train, validation and test\n",
"train_set, val_set, test_set = split_dataset(\n",
" dataset,\n",
" train_size=args.train_size,\n",
" val_size=args.val_size,\n",
" test_size=args.test_size,\n",
" random_state=args.random_state\n",
")\n",
"\n",
"\n",
"# scale datasets\n",
"train_transformed, val_transformed, test_transformed = scale_dataset(\n",
" train_set, \n",
" val_set, \n",
" test_set,\n",
" args.target_col\n",
")\n",
"\n",
"print(\"Saving data\")\n",
"train_transformed.write_csv(train_data_path)\n",
"val_transformed.write_csv(val_data_path)\n",
"test_transformed.write_csv(test_data_path)\n",
"print(f\"Ending pre-processing\")\n",
"print(f\"===========================================================\")"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "ac9817d5-4915-401e-823b-6a269de58953",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"sklearn_processor = SKLearnProcessor(\n",
" framework_version='1.0-1',\n",
" role=role_arn,\n",
" instance_type=processing_instance_type,\n",
" instance_count=processing_instance_count\n",
")"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "a87b5c9f-f67b-48e8-a49a-4c32b29b8206",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# Optional step\n",
"# Uncomment the delete line below to delete all file in the S3 prefix before begining preprocessing. \n",
"# This is to prevent duplication of data when running this workshop multiple time.\n",
"\n",
"s3 = boto3.resource('s3')\n",
"print(bucket)\n",
"bucket_obj = s3.Bucket(bucket)\n",
"print(f\"{s3_prefix}/data/sm_processed/\")\n",
"files = bucket_obj.objects.filter(Prefix=f\"{s3_prefix}/data/sm_processed/\")\n",
"\n",
"#Uncomment the below line \n",
"files.delete()"
]
},
{
"cell_type": "markdown",
"id": "690cbc5d-52e3-42d3-8a3b-b8a38c26264d",
"metadata": {},
"source": [
"We're now ready to run the Processing job for spliting and scaling the data."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "90766c5d-fc6e-43f3-99ac-847655010f29",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"train_s3_destination = f'{output_path}/train'\n",
"val_s3_destination = f'{output_path}/validation'\n",
"test_s3_destination = f'{output_path}/test'"
]
},
{
"cell_type": "markdown",
"id": "c3c40b76-7add-4de5-8664-0c473993bff5",
"metadata": {},
"source": [
"Let's download the test set to a local directory. We will be using this in notebook 3 to test our deployment"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "86208bf6-cbe4-4e36-a195-9c0a4a159f4f",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"test_s3_destination"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "c1fb6240-f3cd-4fd8-b189-57f72ea7059e",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from IPython.core.display import display, HTML\n",
"# code=can be a s3 uri for the input script\n",
"job_name = f\"processing-{strftime('%Y-%m-%d-%H-%M-%S')}\"\n",
"\n",
"display(\n",
" HTML(\n",
" 'Review Processing Job'.format(\n",
" region, job_name\n",
" )\n",
" )\n",
")\n",
"sklearn_processor.run(\n",
" code='pipeline_scripts/preprocessing/script.py',\n",
" job_name=job_name,\n",
" outputs=[\n",
" ProcessingOutput(\n",
" output_name='train',\n",
" destination=train_s3_destination,\n",
" source='/opt/ml/processing/output/train'\n",
" ), \n",
" ProcessingOutput(\n",
" output_name='validation',\n",
" destination=val_s3_destination,\n",
" source='/opt/ml/processing/output/validation'\n",
" ),\n",
" ProcessingOutput(\n",
" output_name='test',\n",
" destination=test_s3_destination,\n",
" source='/opt/ml/processing/output/test'\n",
" )\n",
" ],\n",
" # notice that all arguments passed to a SageMaker processing job should be strings as they are transformed to command line parameters.\n",
" # Your read_parameters function will handle the data types for your code \n",
" arguments=[\n",
" \"--feature_group_name\", feature_group_name,\n",
" \"--train_size\", str(train_size),\n",
" \"--val_size\", str(val_size),\n",
" \"--test_size\", str(test_size),\n",
" \"--random_state\", str(random_seed),\n",
" \"--region\", region\n",
" ]\n",
")\n",
"preprocessing_job_description = sklearn_processor.jobs[-1].describe()"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "111c5d55-a4f5-4fc4-a8d4-0f4088635208",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from sagemaker.s3 import S3Downloader\n",
"\n",
"S3Downloader.download(s3_uri=test_s3_destination, local_path=processed_test_dir)"
]
},
{
"cell_type": "markdown",
"id": "8708f469-dc4c-432e-b93a-30fb00ea4139",
"metadata": {},
"source": [
"### Sanity Check"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "2f533b49-cccf-4411-bb24-3177ea45bfec",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"print(f'{output_path}/train')\n",
"train_ds = ray.data.read_csv(f'{output_path}/train')\n",
"train_ds.count()"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "12e34a36-9a08-455e-a8d1-be75e3e8458d",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%store feature_group_name\n",
"%store s3_prefix\n",
"%store output_path\n",
"%store train_s3_destination\n",
"%store val_s3_destination\n",
"%store test_s3_destination\n",
"%store raw_s3\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "43b5d734-7054-4626-83f2-1c64c564718b",
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"availableInstances": [
{
"_defaultOrder": 0,
"_isFastLaunch": true,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 4,
"name": "ml.t3.medium",
"vcpuNum": 2
},
{
"_defaultOrder": 1,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 8,
"name": "ml.t3.large",
"vcpuNum": 2
},
{
"_defaultOrder": 2,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 16,
"name": "ml.t3.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 3,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 32,
"name": "ml.t3.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 4,
"_isFastLaunch": true,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 8,
"name": "ml.m5.large",
"vcpuNum": 2
},
{
"_defaultOrder": 5,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 16,
"name": "ml.m5.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 6,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 32,
"name": "ml.m5.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 7,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 64,
"name": "ml.m5.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 8,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 128,
"name": "ml.m5.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 9,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 192,
"name": "ml.m5.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 10,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 256,
"name": "ml.m5.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 11,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 384,
"name": "ml.m5.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 12,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 8,
"name": "ml.m5d.large",
"vcpuNum": 2
},
{
"_defaultOrder": 13,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 16,
"name": "ml.m5d.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 14,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 32,
"name": "ml.m5d.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 15,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 64,
"name": "ml.m5d.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 16,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 128,
"name": "ml.m5d.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 17,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 192,
"name": "ml.m5d.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 18,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 256,
"name": "ml.m5d.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 19,
"_isFastLaunch": false,
"category": "General purpose",
"gpuNum": 0,
"memoryGiB": 384,
"name": "ml.m5d.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 20,
"_isFastLaunch": true,
"category": "Compute optimized",
"gpuNum": 0,
"memoryGiB": 4,
"name": "ml.c5.large",
"vcpuNum": 2
},
{
"_defaultOrder": 21,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"memoryGiB": 8,
"name": "ml.c5.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 22,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"memoryGiB": 16,
"name": "ml.c5.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 23,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"memoryGiB": 32,
"name": "ml.c5.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 24,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"memoryGiB": 72,
"name": "ml.c5.9xlarge",
"vcpuNum": 36
},
{
"_defaultOrder": 25,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"memoryGiB": 96,
"name": "ml.c5.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 26,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"memoryGiB": 144,
"name": "ml.c5.18xlarge",
"vcpuNum": 72
},
{
"_defaultOrder": 27,
"_isFastLaunch": false,
"category": "Compute optimized",
"gpuNum": 0,
"memoryGiB": 192,
"name": "ml.c5.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 28,
"_isFastLaunch": true,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 16,
"name": "ml.g4dn.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 29,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 32,
"name": "ml.g4dn.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 30,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 64,
"name": "ml.g4dn.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 31,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 128,
"name": "ml.g4dn.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 32,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 4,
"memoryGiB": 192,
"name": "ml.g4dn.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 33,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 256,
"name": "ml.g4dn.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 34,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 61,
"name": "ml.p3.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 35,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 4,
"memoryGiB": 244,
"name": "ml.p3.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 36,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 8,
"memoryGiB": 488,
"name": "ml.p3.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 37,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 8,
"memoryGiB": 768,
"name": "ml.p3dn.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 38,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"memoryGiB": 16,
"name": "ml.r5.large",
"vcpuNum": 2
},
{
"_defaultOrder": 39,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"memoryGiB": 32,
"name": "ml.r5.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 40,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"memoryGiB": 64,
"name": "ml.r5.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 41,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"memoryGiB": 128,
"name": "ml.r5.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 42,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"memoryGiB": 256,
"name": "ml.r5.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 43,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"memoryGiB": 384,
"name": "ml.r5.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 44,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"memoryGiB": 512,
"name": "ml.r5.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 45,
"_isFastLaunch": false,
"category": "Memory Optimized",
"gpuNum": 0,
"memoryGiB": 768,
"name": "ml.r5.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 46,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 16,
"name": "ml.g5.xlarge",
"vcpuNum": 4
},
{
"_defaultOrder": 47,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 32,
"name": "ml.g5.2xlarge",
"vcpuNum": 8
},
{
"_defaultOrder": 48,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 64,
"name": "ml.g5.4xlarge",
"vcpuNum": 16
},
{
"_defaultOrder": 49,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 128,
"name": "ml.g5.8xlarge",
"vcpuNum": 32
},
{
"_defaultOrder": 50,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 1,
"memoryGiB": 256,
"name": "ml.g5.16xlarge",
"vcpuNum": 64
},
{
"_defaultOrder": 51,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 4,
"memoryGiB": 192,
"name": "ml.g5.12xlarge",
"vcpuNum": 48
},
{
"_defaultOrder": 52,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 4,
"memoryGiB": 384,
"name": "ml.g5.24xlarge",
"vcpuNum": 96
},
{
"_defaultOrder": 53,
"_isFastLaunch": false,
"category": "Accelerated computing",
"gpuNum": 8,
"memoryGiB": 768,
"name": "ml.g5.48xlarge",
"vcpuNum": 192
}
],
"instance_type": "ml.t3.medium",
"kernelspec": {
"display_name": "Python 3 (Data Science 3.0)",
"language": "python",
"name": "python3__SAGEMAKER_INTERNAL__arn:aws:sagemaker:us-east-1:081325390199:image/sagemaker-data-science-310-v1"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.10.6"
}
},
"nbformat": 4,
"nbformat_minor": 5
}