{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# SageMaker Inference Pipeline with Scikit Learn and Linear Learner\n",
"ISO20022 pacs.008 inference pipeline notebook. This notebook uses training dataset to perform model training. It uses SageMaker Linear Learner to train a model. The problem is defined to be a `binary classification` problem of accepting or rejecting a pacs.008 message.\n",
"\n",
"Amazon SageMaker provides a very rich set of [builtin algorithms](https://docs.aws.amazon.com/sagemaker/latest/dg/algorithms-choose.html) for model training and development. This notebook uses [Amazon SageMaker Linear Learner Algorithm](https://docs.aws.amazon.com/sagemaker/latest/dg/linear-learner.html) on training dataset to perform model training. The Amazon SageMaker linear learner algorithm provides a solution for both classification and regression problems. With the SageMaker algorithm, you can simultaneously explore different training objectives and choose the best solution from a validation set. You can also explore a large number of models and choose the best. The best model optimizes either of the following:\n",
"* Continuous objectives, such as mean square error, cross entropy loss, absolute error (regression models).\n",
"* Discrete objectives suited for classification, such as F1 measure, precision, recall, or accuracy (classification models).\n",
"\n",
"ML Model development is an iterative process with several tasks that data scientists go through to produce an effective model that can solve business problem. The process typically involves:\n",
"* Data exploration and analysis\n",
"* Feature engineering\n",
"* Model development\n",
"* Model training and tuning\n",
"* Model deployment\n",
"\n",
"We provide the accompanying notebook [pacs008_xgboost_local.ipynb](./pacs008_xgboost_local.ipynb) which demonstrates data exploration, analysis and feature engineering, focussing on text feature engineering. This notebook uses the results of analysis in [pacs008_xgboost_local.ipynb](./pacs008_xgboost_local.ipynb) to create a feature engineering pipeline using [SageMaker Inference Pipeline](https://docs.aws.amazon.com/sagemaker/latest/dg/inference-pipelines.html).\n",
"\n",
"Here we define the ML problem to be a `binary classification` problem, that of predicting if a pacs.008 XML message with be processed sucessfully or lead to exception process. The predicts `Success` i.e. 1 or `Failure` i.e. 0. \n",
"\n",
"**Feature Engineering** \n",
"\n",
"Data pre-processing and featurizing the dataset by incorporating standard techniques or prior knowledge is a standard mechanism to make dataset meaningful for training. Once data has been pre-processed and transformed, it can be finally used to train an ML model using an algorithm. However, when the trained model is used for processing real time or batch prediction requests, the model receives data in a format which needs to be pre-processed (e.g. featurized) before it can be passed to the algorithm. In this notebook, we will demonstrate how you can build your ML Pipeline leveraging the Sagemaker Scikit-learn container and SageMaker XGBoost algorithm. After a model is trained, we deploy the Pipeline (Data preprocessing and XGBoost) as an **Inference Pipeline** behind a **single Endpoint** for real time inference and for **batch inferences** using Amazon SageMaker Batch Transform.\n",
"\n",
"We use pacs.008 xml element `TEXT` to perform feature engineer i.e featurize text into new numeric features that can be used in making prodictions.\n",
"\n",
"Since we featurize `InstrForNxtAgt` to numeric representations during training, we have to pre-processs to transform text into numeric features before using the trained model to make predictions.\n",
"\n",
"**Inference Pipeline**\n",
"\n",
"The diagram below shows how Amazon SageMaker Inference Pipeline works. It is used to deploy multi-container endpoints.\n",
"\n",
"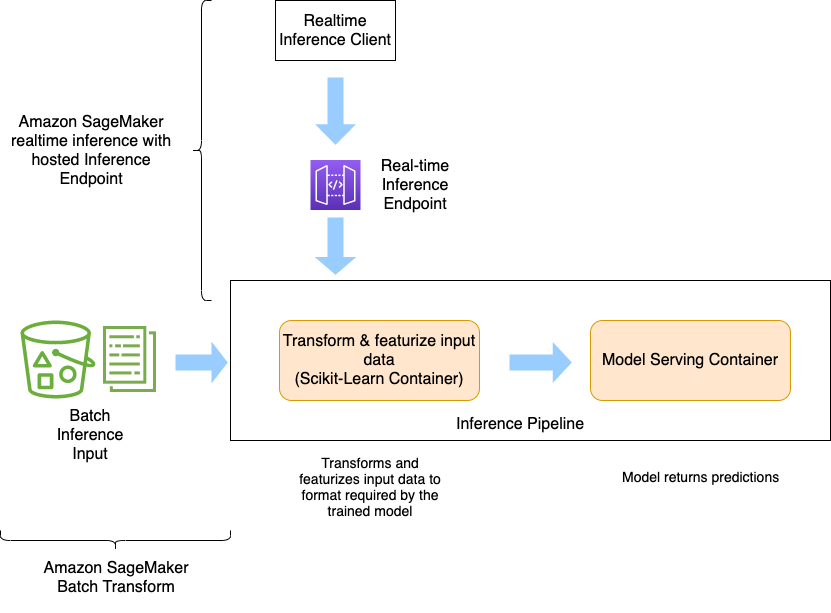\n",
"\n",
"\n",
"**Inference Endpoint** \n",
"\n",
"The diagram below shows the places in the cross-border payment message flow where a call to ML inference endpoint can be injected to get inference from the ML model. The inference result can be used to take additional actions, including corrective actions before sending the message downstream.\n",
"\n",
"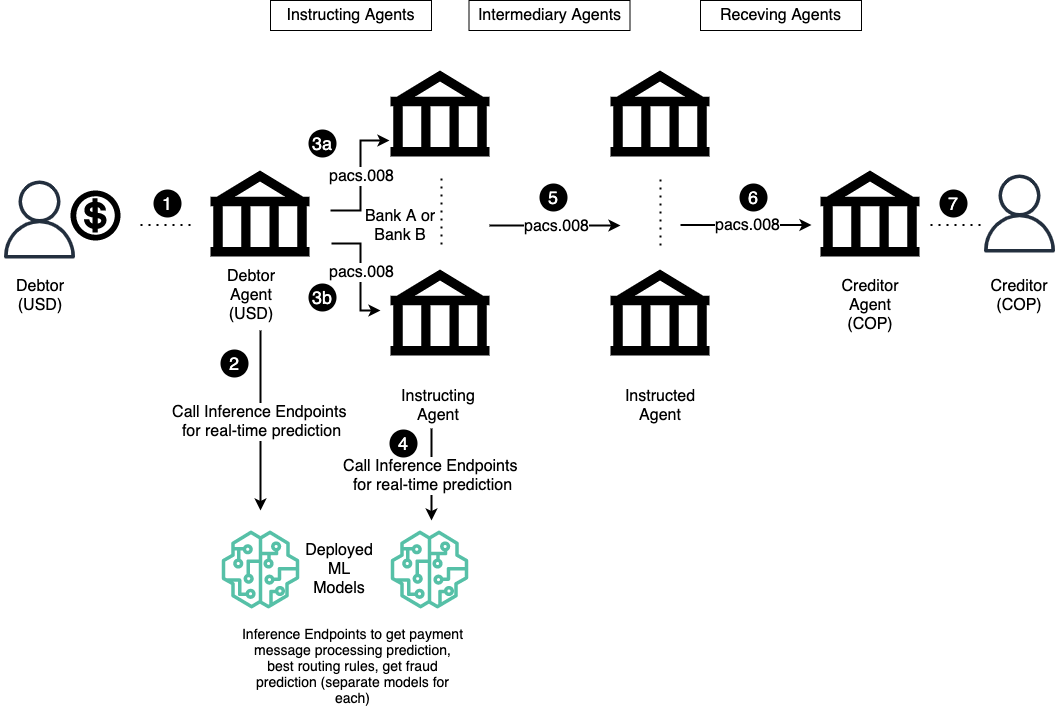\n",
"\n",
"\n",
"**Further Reading:** \n",
"For information on Amazon SageMaker Linear Learner algorithm and SageMaker Inference Pipeline visit the following references: \n",
"\n",
"[SageMaker Linear Learner Algorithm](https://docs.aws.amazon.com/sagemaker/latest/dg/linear-learner.html) \n",
"\n",
"[SageMaker Inference Pipeline](https://docs.aws.amazon.com/sagemaker/latest/dg/inference-pipelines.html)\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Basic Setup\n",
"\n",
"In this step we do basic setup needed for rest of the notebook:\n",
"* Amazon SageMaker API client using boto3\n",
"* Amazon SageMaker session object\n",
"* AWS region\n",
"* AWS IAM role"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import os\n",
"import boto3\n",
"import sagemaker\n",
"from sagemaker import get_execution_role\n",
"\n",
"sm_client = boto3.Session().client('sagemaker')\n",
"sm_session = sagemaker.Session()\n",
"region = boto3.session.Session().region_name\n",
"\n",
"role = get_execution_role()\n",
"print (\"Notebook is running with assumed role {}\".format (role))\n",
"print(\"Working with AWS services in the {} region\".format(region))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Provide S3 Bucket Name"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Working directory for the notebook\n",
"WORKDIR = os.getcwd()\n",
"BASENAME = os.path.dirname(WORKDIR)\n",
"print(f\"WORKDIR: {WORKDIR}\")\n",
"print(f\"BASENAME: {BASENAME}\")\n",
"\n",
"# Create a directory storing local data\n",
"iso20022_data_path = 'iso20022-data'\n",
"if not os.path.exists(iso20022_data_path):\n",
" # Create a new directory because it does not exist \n",
" os.makedirs(iso20022_data_path)\n",
"\n",
"# Store all prototype assets in this bucket\n",
"s3_bucket_name = 'iso20022-prototype-t3'\n",
"s3_bucket_uri = 's3://' + s3_bucket_name\n",
"\n",
"# Prefix for all files in this prototype\n",
"prefix = 'iso20022'\n",
"\n",
"pacs008_prefix = prefix + '/pacs008'\n",
"raw_data_prefix = pacs008_prefix + '/raw-data'\n",
"labeled_data_prefix = pacs008_prefix + '/labeled-data'\n",
"training_data_prefix = pacs008_prefix + '/training-data'\n",
"training_headers_prefix = pacs008_prefix + '/training-headers'\n",
"test_data_prefix = pacs008_prefix + '/test-data'\n",
"training_job_output_prefix = pacs008_prefix + '/training-output'\n",
"\n",
"print(f\"Training data with headers will be uploaded to {s3_bucket_uri + '/' + training_headers_prefix}\")\n",
"print(f\"Training data will be uploaded to {s3_bucket_uri + '/' + training_data_prefix}\")\n",
"print(f\"Test data will be uploaded to {s3_bucket_uri + '/' + test_data_prefix}\")\n",
"print(f\"Training job output will be stored in {s3_bucket_uri + '/' + training_job_output_prefix}\")"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"labeled_data_location = s3_bucket_uri + '/' + labeled_data_prefix\n",
"training_data_w_headers_location = s3_bucket_uri + '/' + training_headers_prefix\n",
"training_data_location = s3_bucket_uri + '/' + training_data_prefix\n",
"test_data_location = s3_bucket_uri + '/' + test_data_prefix\n",
"print(f\"Raw labeled data location = {labeled_data_location}\")\n",
"print(f\"Training data with headers location = {training_data_w_headers_location}\")\n",
"print(f\"Training data location = {training_data_location}\")\n",
"print(f\"Test data location = {test_data_location}\")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Prepare Training Dataset \n",
"\n",
"1. Select training dataset from raw labeled dataset.\n",
"1. Split labeled dataset to training and test datasets."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import numpy as np \n",
"import pandas as pd \n",
"import matplotlib.pyplot as plt\n",
"import seaborn as sns\n",
"import string\n",
"from sklearn.model_selection import train_test_split\n",
"from sklearn import ensemble, metrics, model_selection, naive_bayes\n",
"\n",
"color = sns.color_palette()\n",
"\n",
"%matplotlib inline"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Download raw labeled dataset"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Download labeled raw dataset from S3\n",
"s3_client = boto3.client('s3')\n",
"s3_client.download_file(s3_bucket_name, labeled_data_prefix + '/labeled_data.csv', 'iso20022-data/labeled_data.csv')\n",
"\n",
"# Read the train and test dataset and check the top few lines ##\n",
"labeled_raw_df = pd.read_csv(\"iso20022-data/labeled_data.csv\")"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"labeled_raw_df.head()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Select features for training"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Training features\n",
"fts=[\n",
" 'y_target', \n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_Dbtr_PstlAdr_Ctry', \n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_Cdtr_PstlAdr_Ctry', \n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_RgltryRptg_DbtCdtRptgInd', \n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_RgltryRptg_Authrty_Ctry', \n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_RgltryRptg_Dtls_Cd',\n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_InstrForNxtAgt_InstrInf',\n",
"]\n",
"\n",
"# New data frame with selected features\n",
"selected_df = labeled_raw_df[fts]\n",
" \n",
"selected_df.head()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Rename columns\n",
"selected_df = selected_df.rename(columns={\n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_Dbtr_PstlAdr_Ctry': 'Dbtr_PstlAdr_Ctry',\n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_Cdtr_PstlAdr_Ctry': 'Cdtr_PstlAdr_Ctry',\n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_RgltryRptg_DbtCdtRptgInd': 'RgltryRptg_DbtCdtRptgInd',\n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_RgltryRptg_Authrty_Ctry': 'RgltryRptg_Authrty_Ctry',\n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_RgltryRptg_Dtls_Cd': 'RgltryRptg_Dtls_Cd',\n",
" 'Document_FIToFICstmrCdtTrf_CdtTrfTxInf_InstrForNxtAgt_InstrInf': 'InstrForNxtAgt',\n",
"})\n",
"\n",
"selected_df.head()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from sklearn.preprocessing import LabelEncoder\n",
"\n",
"# Assign Pandas data types.\n",
"categorical_fts=[\n",
" 'Dbtr_PstlAdr_Ctry', \n",
" 'Cdtr_PstlAdr_Ctry',\n",
" 'RgltryRptg_DbtCdtRptgInd', \n",
" 'RgltryRptg_Authrty_Ctry', \n",
" 'RgltryRptg_Dtls_Cd'\n",
"]\n",
"\n",
"integer_fts=[\n",
" \n",
"]\n",
"\n",
"numeric_fts=[\n",
" \n",
"]\n",
"\n",
"text_fts=[\n",
"# Leave text as object \n",
"# 'InstrForNxtAgt'\n",
"]\n",
"\n",
"# Categorical features to categorical data type.\n",
"for col in categorical_fts:\n",
" selected_df[col] = selected_df[col].astype(str).astype('category')\n",
"\n",
"# Integer features to int64 data type. \n",
"for col in integer_fts:\n",
" selected_df[col] = selected_df[col].astype(str).astype('int64')\n",
" \n",
"# Numeric features to float64 data type. \n",
"for col in numeric_fts:\n",
" selected_df[col] = selected_df[col].astype(str).astype('float64')\n",
"\n",
"# Text features to string data type. \n",
"for col in text_fts:\n",
" selected_df[col] = selected_df[col].astype(str).astype('string')\n",
"\n",
"label_encoder = LabelEncoder()\n",
"selected_df['y_target'] = label_encoder.fit_transform(selected_df['y_target'])\n",
" \n",
"selected_df.dtypes"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"selected_df.info()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"selected_df"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X_train_df, X_test_df, y_train_df, y_test_df = train_test_split(selected_df, selected_df['y_target'], test_size=0.20, random_state=299, shuffle=True)\n",
"\n",
"print(\"Number of rows in train dataset : \",X_train_df.shape[0])\n",
"print(\"Number of rows in test dataset : \",X_test_df.shape[0])"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X_train_df"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X_test_df"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"## Save training and test datasets to CSV\n",
"\n",
"train_data_w_headers_output_path = 'iso20022-data/train_data_w_headers.csv'\n",
"print(f'Saving training data with headers to {train_data_w_headers_output_path}')\n",
"X_train_df.to_csv(train_data_w_headers_output_path, index=False)\n",
"\n",
"train_data_output_path = 'iso20022-data/train_data.csv'\n",
"print(f'Saving training data without headers to {train_data_output_path}')\n",
"X_train_df.to_csv(train_data_output_path, header=False, index=False)\n",
"\n",
"test_data_output_path = 'iso20022-data/test_data.csv'\n",
"print(f'Saving test data without headers to {test_data_output_path}')\n",
"X_test_df.to_csv(test_data_output_path, header=False, index=False)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Upload training and test datasets to S3 for training"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"train_input_data_location = sm_session.upload_data(\n",
" path=train_data_w_headers_output_path,\n",
" bucket=s3_bucket_name,\n",
" key_prefix=training_headers_prefix,\n",
")\n",
"print(f'Uploaded traing data with headers to: {train_input_data_location}')\n",
"\n",
"train_input_data_location = sm_session.upload_data(\n",
" path=train_data_output_path,\n",
" bucket=s3_bucket_name,\n",
" key_prefix=training_data_prefix,\n",
")\n",
"print(f'Uploaded data without headers to: {train_input_data_location}')\n",
"\n",
"test_input_data_location = sm_session.upload_data(\n",
" path=test_data_output_path,\n",
" bucket=s3_bucket_name,\n",
" key_prefix=test_data_prefix,\n",
")\n",
"print(f'Uploaded data without headers to: {test_input_data_location}')"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Feature Engineering"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Create a Scikit-learn script to train with \n",
"To run Scikit-learn on Sagemaker `SKLearn` Estimator with a script as an entry point. The training script is very similar to a training script you might run outside of SageMaker, but you can access useful properties about the training environment through various environment variables, such as:\n",
"\n",
"* SM_MODEL_DIR: A string representing the path to the directory to write model artifacts to. These artifacts are uploaded to S3 for model hosting.\n",
"* SM_OUTPUT_DIR: A string representing the filesystem path to write output artifacts to. Output artifacts may include checkpoints, graphs, and other files to save, not including model artifacts. These artifacts are compressed and uploaded to S3 to the same S3 prefix as the model artifacts.\n",
"\n",
"Supposing two input channels, 'train' and 'test', were used in the call to the Chainer estimator's fit() method, the following will be set, following the format SM_CHANNEL_[channel_name]:\n",
"\n",
"* SM_CHANNEL_TRAIN: A string representing the path to the directory containing data in the 'train' channel\n",
"* SM_CHANNEL_TEST: Same as above, but for the 'test' channel.\n",
"\n",
"A typical training script loads data from the input channels, configures training with hyperparameters, trains a model, and saves a model to model_dir so that it can be hosted later. Hyperparameters are passed to your script as arguments and can be retrieved with an argparse.ArgumentParser instance."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Create SageMaker Scikit Estimator \n",
"\n",
"To run our Scikit-learn training script on SageMaker, we construct a `sagemaker.sklearn.estimator.sklearn` estimator, which accepts several constructor arguments:\n",
"\n",
"* __entry_point__: The path to the Python script SageMaker runs for training and prediction.\n",
"* __role__: Role ARN\n",
"* __framework_version__: Scikit-learn version you want to use for executing your model training code.\n",
"* __train_instance_type__ *(optional)*: The type of SageMaker instances for training. __Note__: Because Scikit-learn does not natively support GPU training, Sagemaker Scikit-learn does not currently support training on GPU instance types.\n",
"* __sagemaker_session__ *(optional)*: The session used to train on Sagemaker."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from sagemaker.sklearn.estimator import SKLearn\n",
"\n",
"preprocessing_job_name = 'pacs008-preprocessor-ll'\n",
"print('data preprocessing job name: ' + preprocessing_job_name)\n",
"\n",
"FRAMEWORK_VERSION = \"0.23-1\"\n",
"source_dir = \"../sklearn-transformers\"\n",
"script_file = \"pacs008_sklearn_featurizer.py\"\n",
"\n",
"sklearn_preprocessor = SKLearn(\n",
" entry_point=script_file,\n",
" source_dir=source_dir,\n",
" role=role,\n",
" framework_version=FRAMEWORK_VERSION,\n",
" instance_type=\"ml.c4.xlarge\",\n",
" sagemaker_session=sm_session,\n",
" base_job_name=preprocessing_job_name,\n",
")"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"sklearn_preprocessor.fit({\"train\": train_input_data_location})"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Batch transform our training data \n",
"Now that our proprocessor is properly fitted, let's go ahead and preprocess our training data. Let's use batch transform to directly preprocess the raw data and store right back into s3."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Define a SKLearn Transformer from the trained SKLearn Estimator\n",
"transformer = sklearn_preprocessor.transformer(\n",
" instance_count=1,\n",
" instance_type=\"ml.m5.xlarge\",\n",
" assemble_with=\"Line\",\n",
" accept=\"text/csv\",\n",
")"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Preprocess training input\n",
"transformer.transform(train_input_data_location, content_type=\"text/csv\")\n",
"print(\"Waiting for transform job: \" + transformer.latest_transform_job.job_name)\n",
"transformer.wait()\n",
"preprocessed_train = transformer.output_path"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Train a Linear Learner Model"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Fit a LinearLearner Model with the preprocessed data \n",
"Let's take the preprocessed training data and fit a LinearLearner Model. Sagemaker provides prebuilt algorithm containers that can be used with the Python SDK. The previous Scikit-learn job preprocessed the labeled raw pacs.008 dataset into useable training data that we can now use to fit a binary classifier Linear Learner model.\n",
"\n",
"For more on Linear Learner see: https://docs.aws.amazon.com/sagemaker/latest/dg/linear-learner.html"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from sagemaker.image_uris import retrieve\n",
"\n",
"ll_image = retrieve(\"linear-learner\", boto3.Session().region_name)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Set job name\n",
"training_job_name = 'pacs008-ll-training'\n",
"print('Linear Learner training job name: ' + training_job_name)\n",
"\n",
"# S3 bucket for storing model artifacts\n",
"training_job_output_location = s3_bucket_uri + '/' + training_job_output_prefix + '/ll_model'\n",
"\n",
"ll_estimator = sagemaker.estimator.Estimator(\n",
" ll_image,\n",
" role,\n",
" instance_count=1,\n",
" instance_type=\"ml.m4.2xlarge\",\n",
" volume_size=20,\n",
" max_run=3600,\n",
" input_mode=\"File\",\n",
" output_path=training_job_output_location,\n",
" sagemaker_session=sm_session,\n",
" base_job_name=training_job_name,\n",
")\n",
"\n",
"# binary_classifier_model_selection_criteria: accuracy is default\n",
"# - accuracy | f_beta | precision_at_target_recall |recall_at_target_precision | loss_function\n",
"# feature_dim=auto, # auto or actual number, default is auto\n",
"# epochs=15, default is 15\n",
"# learning_rate=auto or actual number 0.05 or 0.005\n",
"# loss=logistic | auto |hinge_loss, default is logistic\n",
"# mini_batch_size=32, default is 1000\n",
"# num_models=auto, or a number\n",
"# optimizer=auto or sgd | adam | rmsprop\n",
"ll_estimator.set_hyperparameters(\n",
" predictor_type=\"binary_classifier\",\n",
" binary_classifier_model_selection_criteria=\"accuracy\",\n",
" epochs=15,\n",
" mini_batch_size=32)\n",
"\n",
"ll_train_data = sagemaker.inputs.TrainingInput(\n",
" preprocessed_train, # set after preprocessing job completes\n",
" distribution=\"FullyReplicated\",\n",
" content_type=\"text/csv\",\n",
" s3_data_type=\"S3Prefix\",\n",
")\n",
"\n",
"data_channels = {\"train\": ll_train_data}\n",
"ll_estimator.fit(inputs=data_channels, logs=True)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Serial Inference Pipeline with Scikit preprocessor and Linear Learner \n",
"## Set up the inference pipeline \n",
"Setting up a Machine Learning pipeline can be done with the Pipeline Model. This sets up a list of models in a single endpoint. We configure our pipeline model with the fitted Scikit-learn inference model (data preprocessing/feature engineering model) and the fitted Linear Learner model. Deploying the model follows the standard ```deploy``` pattern in the SageMaker Python SDK.\n"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from sagemaker.model import Model\n",
"from sagemaker.pipeline import PipelineModel\n",
"import boto3\n",
"from time import gmtime, strftime\n",
"\n",
"timestamp_prefix = strftime(\"%Y-%m-%d-%H-%M-%S\", gmtime())\n",
"\n",
"# The two SageMaker Models: one for data preprocessing, and second for inference\n",
"scikit_learn_inferencee_model = sklearn_preprocessor.create_model()\n",
"linear_learner_model = ll_estimator.create_model()\n",
"\n",
"model_name = \"pacs008-ll-inference-pipeline-\" + timestamp_prefix\n",
"endpoint_name = \"pacs008-ll-inference-pipeline-ep-\" + timestamp_prefix\n",
"sm_model = PipelineModel(\n",
" name=model_name, role=role, models=[scikit_learn_inferencee_model, linear_learner_model]\n",
")\n",
"\n",
"sm_model.deploy(initial_instance_count=1, instance_type=\"ml.c4.xlarge\", endpoint_name=endpoint_name)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Store Model Name and Endpoint Name in Notebook Magic Store\n",
"\n",
"These notebook magic store values are used in the example batch transform notebook."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%store model_name\n",
"%store endpoint_name"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Make a request to our pipeline endpoint \n",
"\n",
"The diagram below shows the places in the cross-border payment message flow where a call to ML inference endpoint can be injected to get inference from the ML model. The inference result can be used to take additional actions, including corrective actions before sending the message downstream.\n",
"\n",
"\n",
"\n",
"Here we just grab the first line from the test data (you'll notice that the inference python script is very particular about the ordering of the inference request data). The ```ContentType``` field configures the first container, while the ```Accept``` field configures the last container. You can also specify each container's ```Accept``` and ```ContentType``` values using environment variables.\n",
"\n",
"We make our request with the payload in ```'text/csv'``` format, since that is what our script currently supports. If other formats need to be supported, this would have to be added to the ```output_fn()``` method in our entry point. Note that we set the ```Accept``` to ```application/json```, since Linear Learner does not support ```text/csv``` ```Accept```. The inference output in this case is trying to predict `Success` or `Failure` of ISO20022 pacs.008 payment message using only the subset of message XML elements in the message i.e. features on which model was trained. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from sagemaker.predictor import Predictor\n",
"from sagemaker.serializers import CSVSerializer\n",
"\n",
"# payload_1, expect: Failure\n",
"#payload_1 = \"US,GB,,,,/SVC/It is to be delivered in three days. Greater than three days penalty add 2bp per day\"\n",
"payload_1 = \"MX,GB,,,,/SVC/It is to be delivered in four days. Greater than four days penalty add 2bp per day\"\n",
"\n",
"# payload_2, expect: Success\n",
"payload_2 = \"MX,GB,,,,\"\n",
"#payload_2 = \"US,IE,,,,/TRSY/Treasury Services Platinum Customer\"\n",
"\n",
"# payload_3, expect: Failure\n",
"payload_3 = \"TH,US,,,,/SVC/It is to be delivered in four days. Greater than four days penalty add 2bp per day\"\n",
"#payload_3 = \"CA,US,,,,/SVC/It is to be delivered in three days. Greater than three days penalty add 2bp per day\"\n",
"\n",
"# payload_4, expect: Success\n",
"payload_4 = \"IN,CA,DEBT,IN,00.P0006,\"\n",
"\n",
"# payload_5, expect: Success\n",
"payload_5 = \"IE,IN,CRED,IN,0,/REG/15.X0003 FDI in Transportation\"\n",
"# Failure\n",
"payload_5 = \"IE,IN,CRED,IN,0,/REG/15.X0009 FDI in Agriculture \"\n",
"# Failure\n",
"payload_5 = \"IE,IN,CRED,IN,0,/REG/15.X0004 retail\"\n",
"\n",
"# payload_6, expect: Failure\n",
"payload_6 = \"IE,IN,CRED,IN,0,/REG/99.C34698\"\n",
"#payload_6 = \"MX,IE,,,,/TRSY/eweweww\"\n",
"\n",
"endpoint_name = 'pacs008-ll-inference-pipeline-ep-2021-11-25-00-58-52'\n",
"\n",
"predictor = Predictor(\n",
" endpoint_name=endpoint_name, sagemaker_session=sm_session, serializer=CSVSerializer()\n",
")\n",
"\n",
"print(f\"1. Expect Failure i.e. 0, {predictor.predict(payload_1)}\")\n",
"print(f\"2. Expect Success i.e. 1, {predictor.predict(payload_2)}\")\n",
"print(f\"3. Expect Failure i.e. 0, {predictor.predict(payload_3)}\")\n",
"print(f\"4. Expect Success i.e. 1, {predictor.predict(payload_4)}\")\n",
"print(f\"5. Expect Success i.e. 1, {predictor.predict(payload_5)}\")\n",
"print(f\"6. Expect Failure i.e. 0, {predictor.predict(payload_6)}\")\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Delete Endpoint\n",
"Once we are finished with the endpoint, we clean up the resources!"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"sm_client = sm_session.boto_session.client(\"sagemaker\")\n",
"sm_client.delete_endpoint(EndpointName=endpoint_name)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"instance_type": "ml.t3.medium",
"kernelspec": {
"display_name": "Python 3 (Data Science)",
"language": "python",
"name": "python3__SAGEMAKER_INTERNAL__arn:aws:sagemaker:us-east-1:081325390199:image/datascience-1.0"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.10"
}
},
"nbformat": 4,
"nbformat_minor": 5
}