{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# MLOps: Initial - Enable experimentation at scale"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Contents\n",
"\n",
"- [Introduction](#Introduction)\n",
"- [Processing our dataset locally](#Processing-our-dataset-locally)\n",
"- [SageMaker Processing](#SageMaker-Processing)\n",
"- [SageMaker Feature Store](#SageMaker-Feature-Store)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Introduction"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This is our first notebook which will explore the data preparation stage of the ML workflow.\n",
"\n",
"Here, we will put on the hat of `Data Scientist`/`Data Engineer` and will perform the tasks of gathering datasets, pre-processing those datasets to align with our upcoming Training needs. As part of this exercise, we will start by performing these steps manually inside our Notebook local environment. Then we will learn how to bring scale these steps using managed SageMaker processing capabilities. In the last step, we we will save the outcomes of our data processing to a SageMaker Feature Store.\n",
"\n",
"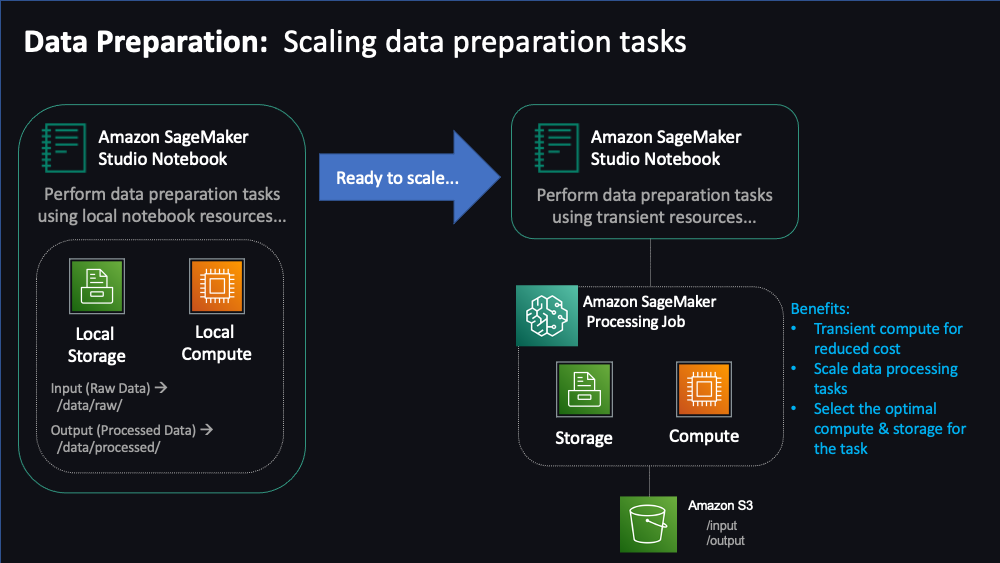"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"To enable you to run these notebooks within a reasonable time (typically less than an hour), the use case is a straightforward regression task: predicting house prices based on a synthetic housing dataset. This dataset contains 8 housing features. Features include year built, number of bedrooms, size of lot, etc...\n",
"\n",
"To begin, we'll import some necessary packages and set up directories for local training and test data. We'll also set up a SageMaker Session to perform various operations, and specify an Amazon S3 bucket to hold input data and output. The default bucket used here is created by SageMaker if it doesn't already exist, and named in accordance with the AWS account ID and AWS Region.\n",
"\n",
"Let's get started!"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Imports**"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"pip install -U sagemaker"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"import sagemaker\n",
"import boto3\n",
"import pandas as pd\n",
"from time import strftime\n",
"from sagemaker.sklearn.processing import SKLearnProcessor\n",
"from sagemaker.processing import ProcessingInput, ProcessingOutput\n",
"from sklearn.model_selection import train_test_split\n",
"from sklearn.preprocessing import StandardScaler\n",
"import os\n",
"import time\n",
"from sagemaker.feature_store.feature_group import FeatureGroup"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Session variables**"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"local_data_path = \"data/raw/house_pricing.csv\"\n",
"\n",
"# setting train, validation and test sizes as strings as required by sagemaker arguments\n",
"train_size = 0.6\n",
"val_size = 0.2\n",
"test_size = 0.2\n",
"random_seed = 42 # setting random seed to ensure compatible results over multiple executions\n",
"\n",
"# Useful SageMaker variables\n",
"sess = sagemaker.Session()\n",
"bucket = sess.default_bucket()\n",
"role_arn= sagemaker.get_execution_role()\n",
"region = sess.boto_region_name\n",
"\n",
"# Local data paths\n",
"pipeline_scripts_dir = os.path.join(os.getcwd(), 'pipeline_scripts')\n",
"os.makedirs(pipeline_scripts_dir, exist_ok=True)\n",
"\n",
"processed_dir = os.path.join(os.getcwd(), 'data/processed')\n",
"os.makedirs(processed_dir, exist_ok=True)\n",
"\n",
"processed_train_dir = os.path.join(os.getcwd(), 'data/processed/train')\n",
"os.makedirs(processed_train_dir, exist_ok=True)\n",
"\n",
"processed_validation_dir = os.path.join(os.getcwd(), 'data/processed/validation')\n",
"os.makedirs(processed_validation_dir, exist_ok=True)\n",
"\n",
"processed_test_dir = os.path.join(os.getcwd(), 'data/processed/test')\n",
"os.makedirs(processed_test_dir, exist_ok=True)\n",
"\n",
"script_processed_dir = os.path.join(os.getcwd(), 'data/script_processed')\n",
"os.makedirs(script_processed_dir, exist_ok=True)\n",
"\n",
"script_processed_train_dir = os.path.join(os.getcwd(), 'data/script_processed/train')\n",
"os.makedirs(script_processed_train_dir, exist_ok=True)\n",
"\n",
"script_processed_validation_dir = os.path.join(os.getcwd(), 'data/script_processed/validation')\n",
"os.makedirs(script_processed_validation_dir, exist_ok=True)\n",
"\n",
"script_processed_test_dir = os.path.join(os.getcwd(), 'data/script_processed/test')\n",
"os.makedirs(script_processed_test_dir, exist_ok=True)\n",
"\n",
"sm_processed_dir = os.path.join(os.getcwd(), 'data/sm_processed')\n",
"os.makedirs(sm_processed_dir, exist_ok=True)\n",
"\n",
"sm_processed_train_dir = os.path.join(os.getcwd(), 'data/sm_processed/train')\n",
"os.makedirs(sm_processed_train_dir, exist_ok=True)\n",
"\n",
"sm_processed_validation_dir = os.path.join(os.getcwd(), 'data/sm_processed/validation')\n",
"os.makedirs(sm_processed_validation_dir, exist_ok=True)\n",
"\n",
"sm_processed_test_dir = os.path.join(os.getcwd(), 'data/sm_processed/test')\n",
"os.makedirs(sm_processed_test_dir, exist_ok=True)\n",
"\n",
"# Data paths in S3\n",
"s3_prefix = 'mlops-workshop'\n",
"\n",
"# SageMaker Processing variables\n",
"processing_instance_type = 'ml.m5.large'\n",
"processing_instance_count = 1\n",
"output_path = f's3://{bucket}/{s3_prefix}/data/sm_processed'"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Processing our dataset locally"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Reading Data**\n",
"\n",
"First we'll read our dataset from the CSV file"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# reading data\n",
"df = pd.read_csv(local_data_path)\n",
"df"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Changing target to the first column in the DataFrame**\n",
"\n",
"Some SageMaker built-in algorithms for CSV training assume that the target variable is in the first column and that the CSV does not have a header record. Even though we'll be bringing our own model, let's do this change so that if we feel like using built-in algorithms, we can easily switch."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# shift column 'PRICE' to first position\n",
"first_column = df.pop('PRICE')\n",
" \n",
"# insert column using insert(position,column_name,\n",
"# first_column) function\n",
"df.insert(0, 'PRICE', first_column)\n",
"df"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Splitting and Scaling data**\n",
"\n",
"Now let's process our data for a Machine Learning model.\n",
"\n",
"First we will split the data into train, validation and test datasets."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# splitting data\n",
"rest_size = 1.0 - train_size\n",
"df_train, df_rest = train_test_split(\n",
" df,\n",
" test_size=rest_size,\n",
" train_size=train_size,\n",
" random_state=random_seed\n",
")\n",
"df_val, df_test = train_test_split(\n",
" df_rest,\n",
" test_size=(test_size / rest_size),\n",
" train_size=(val_size / rest_size),\n",
" random_state=random_seed\n",
")\n",
"df_train.reset_index(inplace=True, drop=True)\n",
"df_val.reset_index(inplace=True, drop=True)\n",
"df_test.reset_index(inplace=True, drop=True)\n",
"train_perc = int((len(df_train)/len(df)) * 100)\n",
"print(f\"Training size: {len(df_train)} - {train_perc}% of total\")\n",
"val_perc = int((len(df_val)/len(df)) * 100)\n",
"print(f\"Val size: {len(df_val)} - {val_perc}% of total\")\n",
"test_perc = int((len(df_test)/len(df)) * 100)\n",
"print(f\"Test size: {len(df_test)} - {test_perc}% of total\")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Next we scale the data based on the training dataset"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# scaling data\n",
"scaler_data = StandardScaler()\n",
" \n",
"# fit scaler to training dataset\n",
"print(\"Fitting scaling to training data and transforming dataset...\")\n",
"df_train_transformed = pd.DataFrame(\n",
" scaler_data.fit_transform(df_train), \n",
" columns=df_train.columns\n",
")\n",
"df_train_transformed['PRICE'] = df_train['PRICE']\n",
"df_train_transformed.head()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"and apply this scaling to the validation"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# apply scaler to validation dataset\n",
"print(\"Transforming validation dataset...\")\n",
"df_val_transformed = pd.DataFrame(\n",
" scaler_data.transform(df_val), \n",
" columns=df_val.columns\n",
")\n",
"df_val_transformed['PRICE'] = df_val['PRICE']\n",
"df_val_transformed.head()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"and test datasets"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# apply scaler to test dataset\n",
"print(\"Transforming test dataset...\")\n",
"df_test_transformed = pd.DataFrame(\n",
" scaler_data.transform(df_test), \n",
" columns=df_test.columns\n",
")\n",
"df_test_transformed['PRICE'] = df_test['PRICE']\n",
"df_test_transformed.head()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Awesome. Let's go ahead and save our generated dataset so that we can do some preprocessing locally on our notebook's instance."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# Save data locally\n",
"df_train_transformed.to_csv(processed_train_dir+'/train.csv', sep=',', index=False, header=False)\n",
"df_val_transformed.to_csv(processed_validation_dir+'/validation.csv', sep=',', index=False, header=False)\n",
"df_test_transformed.to_csv(processed_test_dir+'/test.csv', sep=',', index=False, header=False)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now that we've verified our processing code works, let's write it out to a file and apply it against our locally saved raw data that we generated in the cells above."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile ./local_preprocessing.py\n",
"\n",
"import pandas as pd\n",
"import argparse\n",
"import os\n",
"from sklearn.preprocessing import StandardScaler\n",
"from sklearn.model_selection import train_test_split\n",
"\n",
"\n",
"def read_parameters():\n",
" \"\"\"\n",
" Read job parameters\n",
" Returns:\n",
" (Namespace): read parameters\n",
" \"\"\"\n",
" parser = argparse.ArgumentParser()\n",
" parser.add_argument('--train_size', type=float, default=0.6)\n",
" parser.add_argument('--val_size', type=float, default=0.2)\n",
" parser.add_argument('--test_size', type=float, default=0.2)\n",
" parser.add_argument('--random_state', type=int, default=42)\n",
" parser.add_argument('--target_col', type=str, default='PRICE')\n",
" params, _ = parser.parse_known_args()\n",
" return params\n",
"\n",
"\n",
"def change_target_to_first_col(df, target_col):\n",
" # shift column 'PRICE' to first position\n",
" first_column = df.pop(target_col)\n",
" \n",
" # insert column using insert(position,column_name,\n",
" # first_column) function\n",
" df.insert(0, target_col, first_column)\n",
" return df\n",
"\n",
"\n",
"def split_dataset(df, train_size, val_size, test_size, random_state=None):\n",
" \"\"\"\n",
" Split dataset into train, validation and test samples\n",
" Args:\n",
" df (pandas.DataFrame): input data\n",
" train_size (float): ratio of data to use as training dataset\n",
" val_size (float): ratio of data to use as validation dataset\n",
" test_size (float): ratio of data to use as test dataset\n",
" random_state (int): Pass an int for reproducible output across multiple function calls.\n",
" Returns:\n",
" df_train (pandas.DataFrame): train dataset\n",
" df_val (pandas.DataFrame): validation dataset\n",
" df_test (pandas.DataFrame): test dataset\n",
" \"\"\"\n",
" if (train_size + val_size + test_size) != 1.0:\n",
" raise ValueError(\"train_size, val_size and test_size must sum up to 1.0\")\n",
" rest_size = 1 - train_size\n",
" df_train, df_rest = train_test_split(\n",
" df,\n",
" test_size=rest_size,\n",
" train_size=train_size,\n",
" random_state=random_state\n",
" )\n",
" df_val, df_test = train_test_split(\n",
" df_rest,\n",
" test_size=(test_size / rest_size),\n",
" train_size=(val_size / rest_size),\n",
" random_state=random_state\n",
" )\n",
" df_train.reset_index(inplace=True, drop=True)\n",
" df_val.reset_index(inplace=True, drop=True)\n",
" df_test.reset_index(inplace=True, drop=True)\n",
" train_perc = int((len(df_train)/len(df)) * 100)\n",
" print(f\"Training size: {len(df_train)} - {train_perc}% of total\")\n",
" val_perc = int((len(df_val)/len(df)) * 100)\n",
" print(f\"Val size: {len(df_val)} - {val_perc}% of total\")\n",
" test_perc = int((len(df_test)/len(df)) * 100)\n",
" print(f\"Test size: {len(df_test)} - {test_perc}% of total\")\n",
" return df_train, df_val, df_test\n",
"\n",
"\n",
"def scale_dataset(df_train, df_val, df_test, target_col):\n",
" \"\"\"\n",
" Fit StandardScaler to df_train and apply it to df_val and df_test\n",
" Args:\n",
" df_train (pandas.DataFrame): train dataset\n",
" df_val (pandas.DataFrame): validation dataset\n",
" df_test (pandas.DataFrame): test dataset\n",
" target_col (str): target col\n",
" Returns:\n",
" df_train_transformed (pandas.DataFrame): train data scaled\n",
" df_val_transformed (pandas.DataFrame): val data scaled\n",
" df_test_transformed (pandas.DataFrame): test data scaled\n",
" \"\"\"\n",
" scaler_data = StandardScaler()\n",
" \n",
" # fit scaler to training dataset\n",
" print(\"Fitting scaling to training data and transforming dataset...\")\n",
" df_train_transformed = pd.DataFrame(\n",
" scaler_data.fit_transform(df_train), \n",
" columns=df_train.columns\n",
" )\n",
" df_train_transformed[target_col] = df_train[target_col]\n",
" \n",
" # apply scaler to validation and test datasets\n",
" print(\"Transforming validation and test datasets...\")\n",
" df_val_transformed = pd.DataFrame(\n",
" scaler_data.transform(df_val), \n",
" columns=df_val.columns\n",
" )\n",
" df_val_transformed[target_col] = df_val[target_col]\n",
" df_test_transformed = pd.DataFrame(\n",
" scaler_data.transform(df_test), \n",
" columns=df_test.columns\n",
" )\n",
" df_test_transformed[target_col] = df_test[target_col]\n",
" return df_train_transformed, df_val_transformed, df_test_transformed\n",
"\n",
"\n",
"print(f\"===========================================================\")\n",
"print(f\"Starting pre-processing\")\n",
"print(f\"Reading parameters\")\n",
"\n",
"# reading job parameters\n",
"args = read_parameters()\n",
"print(f\"Parameters read: {args}\")\n",
"\n",
"# set input path\n",
"input_data_path = \"data/raw/house_pricing.csv\"\n",
"\n",
"# read data input\n",
"df = pd.read_csv(input_data_path)\n",
"\n",
"# move target to first col\n",
"df = change_target_to_first_col(df, args.target_col)\n",
"\n",
"# split dataset into train, validation and test\n",
"df_train, df_val, df_test = split_dataset(\n",
" df,\n",
" train_size=args.train_size,\n",
" val_size=args.val_size,\n",
" test_size=args.test_size,\n",
" random_state=args.random_state\n",
")\n",
"\n",
"# scale datasets\n",
"df_train_transformed, df_val_transformed, df_test_transformed = scale_dataset(\n",
" df_train, \n",
" df_val, \n",
" df_test,\n",
" args.target_col\n",
")\n",
"\n",
"df_train_transformed.to_csv('data/script_processed/train/train.csv', sep=',', index=False, header=False)\n",
"df_val_transformed.to_csv('data/script_processed/validation/validation.csv', sep=',', index=False, header=False)\n",
"df_test_transformed.to_csv('data/script_processed/test/test.csv', sep=',', index=False, header=False)\n",
"\n",
"\n",
"\n",
"print(f\"Ending pre-processing\")\n",
"print(f\"===========================================================\")"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!python local_preprocessing.py --train_size 0.6 --val_size 0.2 --test_size 0.2 --random_state 42"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Let's compare the `train.csv` file that was an output of the script to our `df_train_transformed` variable we have saved in memory to make sure they're the same and our preprocessing script worked."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from pandas.testing import assert_frame_equal\n",
"locally_processed_df_train_transformed = pd.read_csv(\n",
" 'data/script_processed/train/train.csv', \n",
" sep=',', \n",
" header=None\n",
")\n",
"locally_processed_df_train_transformed.columns = df_train_transformed.columns\n",
"try:\n",
" assert_frame_equal(df_train_transformed, locally_processed_df_train_transformed)\n",
" print(\"Nice! They match.\")\n",
"except Exception as e:\n",
" print(e)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Ok, so you can process data locally, but this is a smaller dataset. What if you need to process hundreds of gigabytes or even terabytes of data? The processing done so far has been constrained by local resources; this notebook is being run on a single instance type that has memory and compute contraints so we can only process so much data with it.\n",
"\n",
"In order to process larger amounts of data in a reasonable time, we really need to distribute our processing across a cluster of instances. Fortunately, SageMaker has a feature called SageMaker Processing that can help us with this task."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## SageMaker Processing\n",
" \n",
"To process large amounts of data, we fortunately will not need to write distributed code oursleves. Instead, we can use [SageMaker Processing](https://docs.aws.amazon.com/sagemaker/latest/dg/processing-job.html) which will do all the processing _outside_ of this notebook's resources and will apply our processing script to multiple data files in parallel.\n",
" \n",
"Keep in mind that in a typical SageMaker workflow, notebooks are only used for initial model development activities and can be run on relatively inexpensive and less powerful instances. However, to run similar tasks at scale, data scientists require access to more powerful SageMaker managed compute instances for data preparation, training, and model hosting tasks. \n",
"\n",
"SageMaker Processing includes off-the-shelf support for [scikit-learn](https://docs.aws.amazon.com/sagemaker/latest/dg/use-scikit-learn-processing-container.html), [PySpark](https://docs.aws.amazon.com/sagemaker/latest/dg/use-spark-processing-container.html), and [other frameworks](https://docs.aws.amazon.com/sagemaker/latest/dg/processing-job-frameworks.html) like Hugging Face, MXNet, PyTorch, TensorFlow, and XGBoost. You can even a Bring Your Own Container if one our our built-in containers does not suit your use case."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"To leverage SageMaker Processing, we'll need our raw data in S3."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# Upload raw data to S3\n",
"raw_data_s3_prefix = '{}/data/raw'.format(s3_prefix)\n",
"raw_s3 = sess.upload_data(path='./data/raw/house_pricing.csv', key_prefix=raw_data_s3_prefix)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Next, we'll simply supply our Python processing script with a simple modifications to replace the local path we saved our processed data to with the correct container path."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile ./pipeline_scripts/preprocessing.py\n",
"\n",
"import pandas as pd\n",
"import argparse\n",
"import os\n",
"from sklearn.preprocessing import StandardScaler\n",
"from sklearn.model_selection import train_test_split\n",
"\n",
"\n",
"def read_parameters():\n",
" \"\"\"\n",
" Read job parameters\n",
" Returns:\n",
" (Namespace): read parameters\n",
" \"\"\"\n",
" parser = argparse.ArgumentParser()\n",
" parser.add_argument('--train_size', type=float, default=0.6)\n",
" parser.add_argument('--val_size', type=float, default=0.2)\n",
" parser.add_argument('--test_size', type=float, default=0.2)\n",
" parser.add_argument('--random_state', type=int, default=42)\n",
" parser.add_argument('--target_col', type=str, default='PRICE')\n",
" params, _ = parser.parse_known_args()\n",
" return params\n",
"\n",
"\n",
"def change_target_to_first_col(df, target_col):\n",
" # shift column 'PRICE' to first position\n",
" first_column = df.pop(target_col)\n",
" \n",
" # insert column using insert(position,column_name,\n",
" # first_column) function\n",
" df.insert(0, target_col, first_column)\n",
" return df\n",
"\n",
"\n",
"def split_dataset(df, train_size, val_size, test_size, random_state=None):\n",
" \"\"\"\n",
" Split dataset into train, validation and test samples\n",
" Args:\n",
" df (pandas.DataFrame): input data\n",
" train_size (float): ratio of data to use as training dataset\n",
" val_size (float): ratio of data to use as validation dataset\n",
" test_size (float): ratio of data to use as test dataset\n",
" random_state (int): Pass an int for reproducible output across multiple function calls.\n",
" Returns:\n",
" df_train (pandas.DataFrame): train dataset\n",
" df_val (pandas.DataFrame): validation dataset\n",
" df_test (pandas.DataFrame): test dataset\n",
" \"\"\"\n",
" if (train_size + val_size + test_size) != 1.0:\n",
" raise ValueError(\"train_size, val_size and test_size must sum up to 1.0\")\n",
" rest_size = 1 - train_size\n",
" df_train, df_rest = train_test_split(\n",
" df,\n",
" test_size=rest_size,\n",
" train_size=train_size,\n",
" random_state=random_state\n",
" )\n",
" df_val, df_test = train_test_split(\n",
" df_rest,\n",
" test_size=(test_size / rest_size),\n",
" train_size=(val_size / rest_size),\n",
" random_state=random_state\n",
" )\n",
" df_train.reset_index(inplace=True, drop=True)\n",
" df_val.reset_index(inplace=True, drop=True)\n",
" df_test.reset_index(inplace=True, drop=True)\n",
" train_perc = int((len(df_train)/len(df)) * 100)\n",
" print(f\"Training size: {len(df_train)} - {train_perc}% of total\")\n",
" val_perc = int((len(df_val)/len(df)) * 100)\n",
" print(f\"Val size: {len(df_val)} - {val_perc}% of total\")\n",
" test_perc = int((len(df_test)/len(df)) * 100)\n",
" print(f\"Test size: {len(df_test)} - {test_perc}% of total\")\n",
" return df_train, df_val, df_test\n",
"\n",
"\n",
"def scale_dataset(df_train, df_val, df_test, target_col):\n",
" \"\"\"\n",
" Fit StandardScaler to df_train and apply it to df_val and df_test\n",
" Args:\n",
" df_train (pandas.DataFrame): train dataset\n",
" df_val (pandas.DataFrame): validation dataset\n",
" df_test (pandas.DataFrame): test dataset\n",
" target_col (str): target col\n",
" Returns:\n",
" df_train_transformed (pandas.DataFrame): train data scaled\n",
" df_val_transformed (pandas.DataFrame): val data scaled\n",
" df_test_transformed (pandas.DataFrame): test data scaled\n",
" \"\"\"\n",
" scaler_data = StandardScaler()\n",
" \n",
" # fit scaler to training dataset\n",
" print(\"Fitting scaling to training data and transforming dataset...\")\n",
" df_train_transformed = pd.DataFrame(\n",
" scaler_data.fit_transform(df_train), \n",
" columns=df_train.columns\n",
" )\n",
" df_train_transformed[target_col] = df_train[target_col]\n",
" \n",
" # apply scaler to validation and test datasets\n",
" print(\"Transforming validation and test datasets...\")\n",
" df_val_transformed = pd.DataFrame(\n",
" scaler_data.transform(df_val), \n",
" columns=df_val.columns\n",
" )\n",
" df_val_transformed[target_col] = df_val[target_col]\n",
" df_test_transformed = pd.DataFrame(\n",
" scaler_data.transform(df_test), \n",
" columns=df_test.columns\n",
" )\n",
" df_test_transformed[target_col] = df_test[target_col]\n",
" return df_train_transformed, df_val_transformed, df_test_transformed\n",
"\n",
"\n",
"print(f\"===========================================================\")\n",
"print(f\"Starting pre-processing\")\n",
"print(f\"Reading parameters\")\n",
"\n",
"# reading job parameters\n",
"args = read_parameters()\n",
"print(f\"Parameters read: {args}\")\n",
"\n",
"# set input and output paths\n",
"input_data_path = \"/opt/ml/processing/input/house_pricing.csv\"\n",
"train_data_path = \"/opt/ml/processing/output/train\"\n",
"val_data_path = \"/opt/ml/processing/output/validation\"\n",
"test_data_path = \"/opt/ml/processing/output/test\"\n",
"\n",
"try:\n",
" os.makedirs(train_data_path)\n",
" os.makedirs(val_data_path)\n",
" os.makedirs(test_data_path)\n",
"except:\n",
" pass\n",
"\n",
"\n",
"# read data input\n",
"df = pd.read_csv(input_data_path)\n",
"\n",
"# move target to first col\n",
"df = change_target_to_first_col(df, args.target_col)\n",
"\n",
"# split dataset into train, validation and test\n",
"df_train, df_val, df_test = split_dataset(\n",
" df,\n",
" train_size=args.train_size,\n",
" val_size=args.val_size,\n",
" test_size=args.test_size,\n",
" random_state=args.random_state\n",
")\n",
"\n",
"# scale datasets\n",
"df_train_transformed, df_val_transformed, df_test_transformed = scale_dataset(\n",
" df_train, \n",
" df_val, \n",
" df_test,\n",
" args.target_col\n",
")\n",
"\n",
"print(\"Saving data\")\n",
"df_train_transformed.to_csv(train_data_path+'/train.csv', sep=',', index=False, header=False)\n",
"df_val_transformed.to_csv(val_data_path+'/validation.csv', sep=',', index=False, header=False)\n",
"df_test_transformed.to_csv(test_data_path+'/test.csv', sep=',', index=False, header=False)\n",
"\n",
"\n",
"\n",
"print(f\"Ending pre-processing\")\n",
"print(f\"===========================================================\")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Since we're using scikit-learn in our processing script, we'll tell SageMaker Processing that we'll need the scikit-learn processor (the container associated with this processor also includes common libraries like `pandas`) which the SageMaker SDK calls `SKLearnProcessor`. This object allows you to specify the instance type to use in the job as well as how many instances you want in your cluster. Although the synthetic housing dataset is quite small, we'll use two instances to showcase how easy it is to spin up a cluster for SageMaker Processing and parallelize your processing code."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"sklearn_processor = SKLearnProcessor(\n",
" framework_version='1.0-1',\n",
" role=role_arn,\n",
" instance_type=processing_instance_type,\n",
" instance_count=processing_instance_count\n",
")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We're now ready to run the Processing job."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from IPython.core.display import display, HTML\n",
"# code=can be a s3 uri for the input script\n",
"job_name = f\"processing-{strftime('%Y-%m-%d-%H-%M-%S')}\"\n",
"\n",
"display(\n",
" HTML(\n",
" 'Review Processing Job'.format(\n",
" region, job_name\n",
" )\n",
" )\n",
")\n",
"sklearn_processor.run(\n",
" code='pipeline_scripts/preprocessing.py',\n",
" job_name=job_name,\n",
" inputs=[\n",
" ProcessingInput(\n",
" source=raw_s3,\n",
" destination='/opt/ml/processing/input',\n",
" s3_data_distribution_type='ShardedByS3Key'\n",
" )\n",
" ],\n",
" outputs=[\n",
" ProcessingOutput(\n",
" output_name='train',\n",
" destination=f'{output_path}/train',\n",
" source='/opt/ml/processing/output/train'\n",
" ), \n",
" ProcessingOutput(\n",
" output_name='validation',\n",
" destination=f'{output_path}/validation',\n",
" source='/opt/ml/processing/output/validation'\n",
" ),\n",
" ProcessingOutput(\n",
" output_name='test',\n",
" destination=f'{output_path}/test',\n",
" source='/opt/ml/processing/output/test'\n",
" )\n",
" ],\n",
" # notice that all arguments passed to a SageMaker processing job should be strings as they are transformed to command line parameters.\n",
" # Your read_parameters function will handle the data types for your code \n",
" arguments=[\n",
" \"--train_size\", str(train_size),\n",
" \"--val_size\", str(val_size),\n",
" \"--test_size\", str(test_size),\n",
" \"--random_state\", str(random_seed)\n",
" ]\n",
")\n",
"preprocessing_job_description = sklearn_processor.jobs[-1].describe()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now that the SageMaker Processing job has finished, it has output the processed data in S3. Let's download that data locally and ensure it's what we expect."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# Download processed data from S3 to local storage\n",
"!aws s3 cp {output_path} ./data/sm_processed/ --recursive"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"sm_processed_df_train_transformed = pd.read_csv('data/sm_processed/train/train.csv', sep=',', header=None)\n",
"sm_processed_df_train_transformed.columns = df_train_transformed.columns\n",
"try:\n",
" assert_frame_equal(df_train_transformed, sm_processed_df_train_transformed)\n",
" print(\"Again they match!\")\n",
"except Exception as e:\n",
" print(e)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## SageMaker Feature Store\n",
" \n",
"Features are inputs to ML models used during training and inference. Features are used repeatedly by multiple teams and feature quality is critical to ensure a highly accurate model. Also, when features used to train models offline in batch are made available for real-time inference, it’s hard to keep the two feature stores synchronized. [SageMaker Feature Store](https://docs.aws.amazon.com/sagemaker/latest/dg/feature-store.html) provides a secured and unified store for feature use across the ML lifecycle. \n",
"\n",
"Let's now exchange the storage of our processed data from s3 to SageMaker Feature Store.\n",
"\n",
"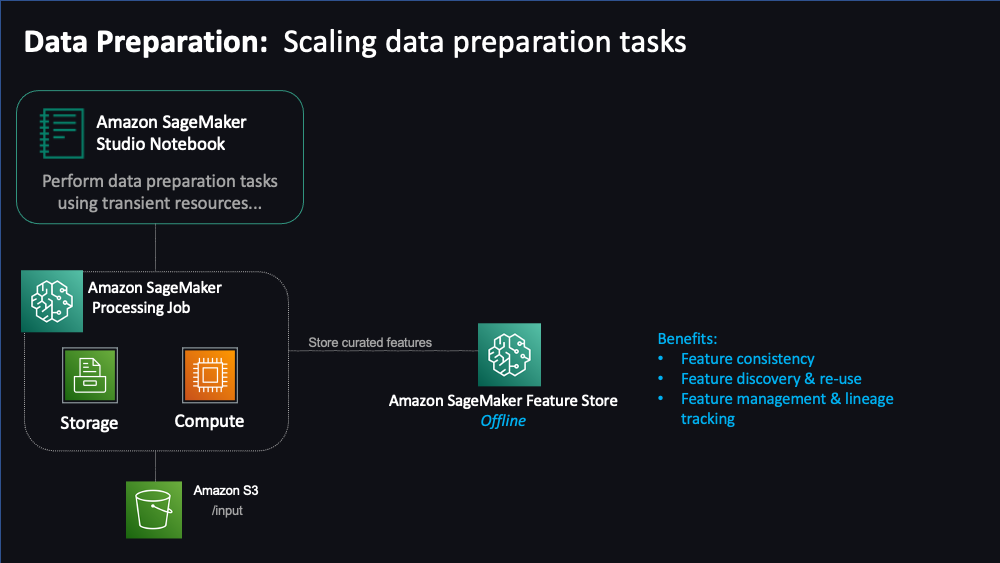\n",
"\n",
"**Feature Groups**\n",
"\n",
"First let's define some feature groups for train, validation and test datasets and a s3 bucket prefix to store your feature store results"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"time_str = '-' + time.strftime('%Y-%m-%d-%H-%M-%S')\n",
"train_feature_group_name = \"fs-train-\"+time_str\n",
"validation_feature_group_name = \"fs-validation-\"+time_str\n",
"test_feature_group_name = \"fs-test-\"+time_str\n",
"bucket_prefix = \"mlops-workshop/feature-store\""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Next, we'll modify our Python processing script to include the creation of feature groups for the train, validation and test datasets and the injection of data to the feature groups."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%%writefile ./pipeline_scripts/preprocessing_with_fs.py\n",
"import subprocess\n",
"import sys\n",
"subprocess.check_call([sys.executable, '-m', 'pip', 'install', 'sagemaker'])\n",
"\n",
"from sagemaker.feature_store.feature_group import FeatureGroup\n",
"import pandas as pd\n",
"import argparse\n",
"import os\n",
"import numpy as np\n",
"from sklearn.preprocessing import StandardScaler\n",
"from sklearn.model_selection import train_test_split\n",
"import time\n",
"import sagemaker\n",
"import boto3\n",
"\n",
"\n",
"def read_parameters():\n",
" \"\"\"\n",
" Read job parameters\n",
" Returns:\n",
" (Namespace): read parameters\n",
" \"\"\"\n",
" parser = argparse.ArgumentParser()\n",
" parser.add_argument('--train_size', type=float, default=0.6)\n",
" parser.add_argument('--val_size', type=float, default=0.2)\n",
" parser.add_argument('--test_size', type=float, default=0.2)\n",
" parser.add_argument('--random_state', type=int, default=42)\n",
" parser.add_argument('--train_feature_group_name', type=str, default='fs-train')\n",
" parser.add_argument('--validation_feature_group_name', type=str, default='fs-validation')\n",
" parser.add_argument('--test_feature_group_name', type=str, default='fs-test')\n",
" parser.add_argument('--bucket_prefix', type=str, default='mlops-workshop/feature-store')\n",
" parser.add_argument('--target_col', type=str, default='PRICE')\n",
" parser.add_argument('--region', type=str)\n",
" parser.add_argument('--role_arn', type=str)\n",
" params, _ = parser.parse_known_args()\n",
" return params\n",
"\n",
"\n",
"def change_target_to_first_col(df, target_col):\n",
" # shift column 'PRICE' to first position\n",
" first_column = df.pop(target_col)\n",
" \n",
" # insert column using insert(position,column_name,\n",
" # first_column) function\n",
" df.insert(0, target_col, first_column)\n",
" return df\n",
"\n",
"\n",
"def split_dataset(df, train_size, val_size, test_size, random_state=None):\n",
" \"\"\"\n",
" Split dataset into train, validation and test samples\n",
" Args:\n",
" df (pandas.DataFrame): input data\n",
" train_size (float): ratio of data to use as training dataset\n",
" val_size (float): ratio of data to use as validation dataset\n",
" test_size (float): ratio of data to use as test dataset\n",
" random_state (int): Pass an int for reproducible output across multiple function calls.\n",
" Returns:\n",
" df_train (pandas.DataFrame): train dataset\n",
" df_val (pandas.DataFrame): validation dataset\n",
" df_test (pandas.DataFrame): test dataset\n",
" \"\"\"\n",
" if (train_size + val_size + test_size) != 1.0:\n",
" raise ValueError(\"train_size, val_size and test_size must sum up to 1.0\")\n",
" rest_size = 1 - train_size\n",
" df_train, df_rest = train_test_split(\n",
" df,\n",
" test_size=rest_size,\n",
" train_size=train_size,\n",
" random_state=random_state\n",
" )\n",
" df_val, df_test = train_test_split(\n",
" df_rest,\n",
" test_size=(test_size / rest_size),\n",
" train_size=(val_size / rest_size),\n",
" random_state=random_state\n",
" )\n",
" df_train.reset_index(inplace=True, drop=True)\n",
" df_val.reset_index(inplace=True, drop=True)\n",
" df_test.reset_index(inplace=True, drop=True)\n",
" train_perc = int((len(df_train)/len(df)) * 100)\n",
" print(f\"Training size: {len(df_train)} - {train_perc}% of total\")\n",
" val_perc = int((len(df_val)/len(df)) * 100)\n",
" print(f\"Val size: {len(df_val)} - {val_perc}% of total\")\n",
" test_perc = int((len(df_test)/len(df)) * 100)\n",
" print(f\"Test size: {len(df_test)} - {test_perc}% of total\")\n",
" return df_train, df_val, df_test\n",
"\n",
"\n",
"def scale_dataset(df_train, df_val, df_test, target_col):\n",
" \"\"\"\n",
" Fit StandardScaler to df_train and apply it to df_val and df_test\n",
" Args:\n",
" df_train (pandas.DataFrame): train dataset\n",
" df_val (pandas.DataFrame): validation dataset\n",
" df_test (pandas.DataFrame): test dataset\n",
" target_col (str): target col\n",
" Returns:\n",
" df_train_transformed (pandas.DataFrame): train data scaled\n",
" df_val_transformed (pandas.DataFrame): val data scaled\n",
" df_test_transformed (pandas.DataFrame): test data scaled\n",
" \"\"\"\n",
" scaler_data = StandardScaler()\n",
" \n",
" # fit scaler to training dataset\n",
" print(\"Fitting scaling to training data and transforming dataset...\")\n",
" df_train_transformed = pd.DataFrame(\n",
" scaler_data.fit_transform(df_train), \n",
" columns=df_train.columns\n",
" )\n",
" df_train_transformed[target_col] = df_train[target_col]\n",
" \n",
" # apply scaler to validation and test datasets\n",
" print(\"Transforming validation and test datasets...\")\n",
" df_val_transformed = pd.DataFrame(\n",
" scaler_data.transform(df_val), \n",
" columns=df_val.columns\n",
" )\n",
" df_val_transformed[target_col] = df_val[target_col]\n",
" df_test_transformed = pd.DataFrame(\n",
" scaler_data.transform(df_test), \n",
" columns=df_test.columns\n",
" )\n",
" df_test_transformed[target_col] = df_test[target_col]\n",
" return df_train_transformed, df_val_transformed, df_test_transformed\n",
"\n",
"\n",
"def prepare_df_for_feature_store(df, data_type):\n",
" \"\"\"\n",
" Add event time and record id to df in order to store it in SageMaker Feature Store\n",
" Args:\n",
" df (pandas.DataFrame): data to be prepared\n",
" data_type (str): train/validation or test\n",
" Returns:\n",
" df (pandas.DataFrame): dataframe with event time and record id\n",
" \"\"\"\n",
" print(f'Preparing {data_type} data for Feature Store..')\n",
" current_time_sec = int(round(time.time()))\n",
" # create event time\n",
" df['event_time'] = pd.Series([current_time_sec]*len(df), dtype=\"float64\")\n",
" # create record id from index\n",
" df['record_id'] = df.reset_index().index\n",
" return df\n",
" \n",
"\n",
"def wait_for_feature_group_creation_complete(feature_group):\n",
" \"\"\"\n",
" Function that waits for feature group to be created in SageMaker Feature Store\n",
" Args:\n",
" feature_group (sagemaker.feature_store.feature_group.FeatureGroup): Feature Group\n",
" \"\"\"\n",
" status = feature_group.describe().get('FeatureGroupStatus')\n",
" print(f'Initial status: {status}')\n",
" while status == 'Creating':\n",
" print(f'Waiting for feature group: {feature_group.name} to be created ...')\n",
" time.sleep(5)\n",
" status = feature_group.describe().get('FeatureGroupStatus')\n",
" if status != 'Created':\n",
" raise SystemExit(f'Failed to create feature group {feature_group.name}: {status}')\n",
" print(f'FeatureGroup {feature_group.name} was successfully created.')\n",
" \n",
"\n",
"def create_feature_group(feature_group_name, sagemaker_session, df, prefix, role_arn):\n",
" \"\"\"\n",
" Create Feature Store Group\n",
" Args:\n",
" feature_group_name (str): Feature Store Group Name\n",
" sagemaker_session (sagemaker.session.Session): sagemaker session\n",
" df (pandas.DataFrame): dataframe to injest used to create features definition\n",
" prefix (str): geature group prefix (train/validation or test)\n",
" role_arn (str): role arn to create feature store\n",
" Returns:\n",
" fs_group (sagemaker.feature_store.feature_group.FeatureGroup): Feature Group\n",
" \"\"\"\n",
" fs_group = FeatureGroup(\n",
" name=feature_group_name, \n",
" sagemaker_session=sagemaker_session\n",
" )\n",
" fs_group.load_feature_definitions(data_frame=df)\n",
" default_bucket = sagemaker_session.default_bucket()\n",
" print(f'Creating feature group: {fs_group.name} ...')\n",
" fs_group.create(\n",
" s3_uri=f's3://{default_bucket}/{prefix}', \n",
" record_identifier_name='record_id', \n",
" event_time_feature_name='event_time', \n",
" role_arn=role_arn, \n",
" enable_online_store=True\n",
" )\n",
" wait_for_feature_group_creation_complete(fs_group)\n",
" return fs_group\n",
"\n",
"\n",
"def ingest_features(fs_group, df):\n",
" \"\"\"\n",
" Ingest features to Feature Store Group\n",
" Args:\n",
" fs_group (sagemaker.feature_store.feature_group.FeatureGroup): Feature Group\n",
" df (pandas.DataFrame): dataframe to injest\n",
" \"\"\"\n",
" print(f'Ingesting data into feature group: {fs_group.name} ...')\n",
" fs_group.ingest(data_frame=df, max_processes=3, wait=True)\n",
" print(f'{len(df)} records ingested into feature group: {fs_group.name}')\n",
" return\n",
"\n",
"\n",
"print(f\"===========================================================\")\n",
"print(f\"Starting pre-processing\")\n",
"print(f\"Reading parameters\")\n",
"\n",
"# reading job parameters\n",
"args = read_parameters()\n",
"print(f\"Parameters read: {args}\")\n",
"sagemaker_session = sagemaker.Session(boto3.Session(region_name=args.region))\n",
"\n",
"# set input path\n",
"input_data_path = \"/opt/ml/processing/input/house_pricing.csv\"\n",
"\n",
"# read data input\n",
"df = pd.read_csv(input_data_path)\n",
"\n",
"# move target to first col\n",
"df = change_target_to_first_col(df, args.target_col)\n",
"\n",
"# split dataset into train, validation and test\n",
"df_train, df_val, df_test = split_dataset(\n",
" df,\n",
" train_size=args.train_size,\n",
" val_size=args.val_size,\n",
" test_size=args.test_size,\n",
" random_state=args.random_state\n",
")\n",
"\n",
"# scale datasets\n",
"df_train_transformed, df_val_transformed, df_test_transformed = scale_dataset(\n",
" df_train, \n",
" df_val, \n",
" df_test,\n",
" args.target_col\n",
")\n",
"\n",
"# prepare datasets for Feature Store\n",
"df_train_transformed_fs = prepare_df_for_feature_store(df_train_transformed, 'train')\n",
"df_val_transformed_fs = prepare_df_for_feature_store(df_val_transformed, 'validation')\n",
"df_test_transformed_fs = prepare_df_for_feature_store(df_test_transformed, 'test')\n",
"\n",
"# injest datasets to Feature Store\n",
"fs_group_train = create_feature_group(\n",
" args.train_feature_group_name, \n",
" sagemaker_session, \n",
" df_train_transformed_fs, \n",
" args.bucket_prefix+'/train',\n",
" args.role_arn\n",
")\n",
"ingest_features(fs_group_train, df_train_transformed_fs)\n",
"\n",
"fs_group_validation = create_feature_group(\n",
" args.validation_feature_group_name, \n",
" sagemaker_session, \n",
" df_val_transformed_fs, \n",
" args.bucket_prefix+'/validation',\n",
" args.role_arn\n",
")\n",
"ingest_features(fs_group_validation, df_val_transformed_fs)\n",
"\n",
"fs_group_test = create_feature_group(\n",
" args.test_feature_group_name, \n",
" sagemaker_session, \n",
" df_test_transformed_fs, \n",
" args.bucket_prefix+'/test',\n",
" args.role_arn\n",
")\n",
"ingest_features(fs_group_test, df_test_transformed_fs)\n",
"\n",
"print(f\"Ending pre-processing\")\n",
"print(f\"===========================================================\")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We're now ready to run the Processing job with the feature store code included on it."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# code=can be a s3 uri for the input script\n",
"job_name = f\"processing-with-fs-{strftime('%Y-%m-%d-%H-%M-%S')}\"\n",
"\n",
"display(\n",
" HTML(\n",
" 'Review Feature Store Processing Job'.format(\n",
" region, job_name\n",
" )\n",
" )\n",
")\n",
"\n",
"sklearn_processor.run(\n",
" code='pipeline_scripts/preprocessing_with_fs.py',\n",
" job_name=job_name,\n",
" inputs=[\n",
" ProcessingInput(\n",
" source=raw_s3,\n",
" destination='/opt/ml/processing/input',\n",
" s3_data_distribution_type='ShardedByS3Key'\n",
" )\n",
" ],\n",
" # notice that all arguments passed to a SageMaker processing job should be strings as they are transformed to command line parameters.\n",
" # Your read_parameters function will handle the data types for your code \n",
" arguments=[\n",
" \"--train_size\", str(train_size),\n",
" \"--val_size\", str(val_size),\n",
" \"--test_size\", str(test_size),\n",
" \"--random_state\", str(random_seed),\n",
" \"--train_feature_group_name\", train_feature_group_name,\n",
" \"--validation_feature_group_name\", validation_feature_group_name,\n",
" \"--test_feature_group_name\", test_feature_group_name,\n",
" \"--bucket_prefix\", bucket_prefix,\n",
" \"--role_arn\", role_arn,\n",
" \"--region\", region\n",
" ]\n",
")\n",
"\n",
"preprocessing_job_description = sklearn_processor.jobs[-1].describe()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Let's now load the features from feature store"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"fs_group_train = FeatureGroup(name=train_feature_group_name, sagemaker_session=sess) "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"train_query = fs_group_train.athena_query()\n",
"train_table = train_query.table_name"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"And let's display the data in a Dataframe:\n",
"\n",
"**Important**: If your Dataframe is empty, you should wait a bit before training again. It takes a while for Feature Store data to be available in Athena Queries."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"fs_df = pd.DataFrame()\n",
"while len(fs_df) == 0:\n",
" if len(fs_df.columns) > 0:\n",
" time.sleep(120)\n",
" query_string = f'SELECT * FROM \"sagemaker_featurestore\".\"{train_table}\" ORDER BY record_id'\n",
" query_results= 'sagemaker-featurestore'\n",
" output_location = f's3://{bucket}/{query_results}/query_results/'\n",
" train_query.run(query_string=query_string, output_location=output_location)\n",
" train_query.wait()\n",
" fs_df = train_query.as_dataframe()\n",
"fs_df"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Adapting the Athenas Query**\n",
"\n",
"Notice that the data before includes extra information that allows us to audit and monitor the data from Feature Store. \n",
"\n",
"You can change your data to include only the data necessary for your model."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"features_to_select = 'price,year_built,square_feet,num_bedrooms,num_bathrooms,lot_acres,garage_spaces,front_porch,deck'\n",
"query_string = f'SELECT {features_to_select} FROM \"sagemaker_featurestore\".\"{train_table}\" ORDER BY record_id'\n",
"query_results= 'sagemaker-featurestore'\n",
"output_location = f's3://{bucket}/{query_results}/query_results/'\n",
"train_query.run(query_string=query_string, output_location=output_location)\n",
"train_query.wait()\n",
"fs_df = train_query.as_dataframe()\n",
"fs_df"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Next, we'll store some variables that will be used in our second notebook..."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%store train_feature_group_name\n",
"%store validation_feature_group_name\n",
"%store test_feature_group_name\n",
"%store s3_prefix\n",
"%store output_path\n",
"%store features_to_select\n",
"%store sm_processed_train_dir\n",
"%store sm_processed_validation_dir\n",
"%store sm_processed_test_dir\n",
"%store raw_s3"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"instance_type": "ml.t3.medium",
"kernelspec": {
"display_name": "Python 3 (Data Science)",
"language": "python",
"name": "python3__SAGEMAKER_INTERNAL__arn:aws:sagemaker:us-east-1:081325390199:image/datascience-1.0"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.10"
}
},
"nbformat": 4,
"nbformat_minor": 4
}