{
"cells": [
{
"cell_type": "markdown",
"id": "97ee8a43",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"# MLOps: Initial - Enable experimentation at scale\n",
"\n",
" \n",
"\t⚠️ PRE-REQUISITE: Before proceeding with this notebook, please ensure that you have executed the 1-data-prep-feature-store.ipynb Notebook\n",
"
"
]
},
{
"cell_type": "markdown",
"id": "4f3ef1bc",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"## Contents\n",
"\n",
"- [Introduction](#Introduction)\n",
"- [Recap](#Recap)\n",
"- [Experiment tracking](#Experiment-tracking)\n",
"- [SageMaker Training](#SageMaker-Training)\n",
"- [SageMaker Training with Automatic Model Tuning (HPO)](#SageMaker-Training-with-Automatic-Model-Tuning-(HPO))\n",
"- [Model Registry](#Model-Registry)"
]
},
{
"cell_type": "markdown",
"id": "c49b5eb2",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"## Introduction\n",
"\n",
"This is our second notebook which will explore the model training stage of the ML workflow.\n",
"\n",
"Here, we will put on the hat of the `Data Scientist` and will perform the task of modeling which includes training a model, performing hyperparameter tuning, evaluating the model and registering high performing candidate models in a model registry. This task is highly iterative in nature and hence we also need to track our experimentation until we reach desired results.\n",
"\n",
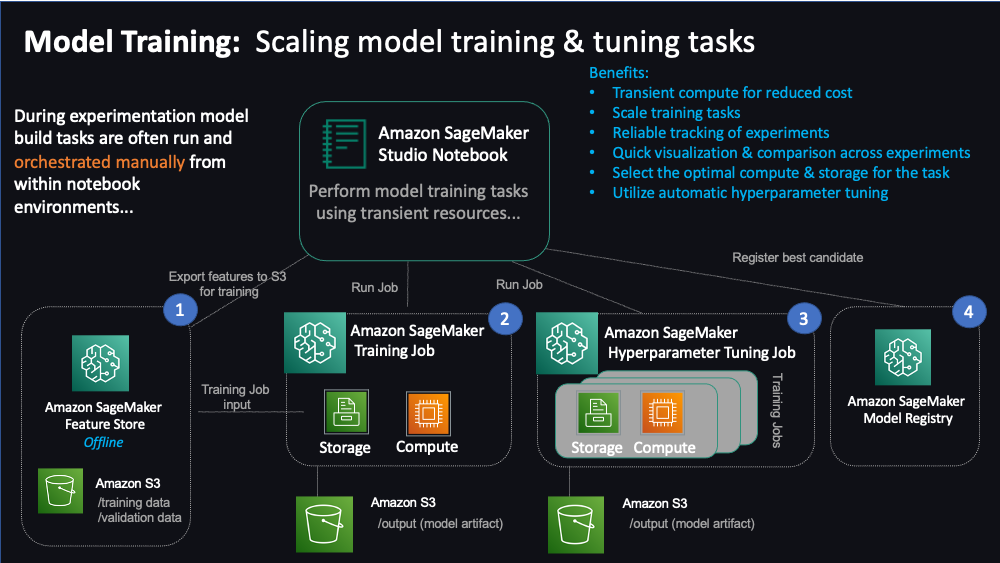
"Similar to the previous notebook on preprocessing datasets, a data scientist could perform the training task using the notebook's local compute & storage using the local data generated during the previous steps. However, this is typically only useful for quick iteration on small datasets. For the purposes of time, we do not cover that option within this notebook. Instead, we'll jump into learning how to bring scale to model development tasks using managed SageMaker training and experiment tracking capabilities combined with curated feature data pulled from SageMaker Feature Store. You'll also perform tuning at scale using SageMaker's automatic hyperparameter tuning capabilities. Then, finally register the best performing model in SageMaker Model Registry. \n",
"\n",
"\n",
"\n",
"\n",
"\n",
"Let's get started!\n",
"\n",
"**Important:** for this example, we will use SageMaker's [XGBoost algorithm](https://docs.aws.amazon.com/sagemaker/latest/dg/xgboost.html) as a built-in model. That means that you don't have to write your model code and SageMaker takes care of it. We will use CSV data as input. For CSV training, the algorithm assumes that the target variable is in the first column and that the CSV does not have a header record. Let's query our Feature Store Group to get the necessary data"
]
},
{
"cell_type": "markdown",
"id": "ab2de56a",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"**Imports**"
]
},
{
"cell_type": "markdown",
"id": "4e493d63",
"metadata": {},
"source": [
"Let's first install the latest version of the SageMaker SDK to ensure we have the current sagemaker-experiments library"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "4491bcf7",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"%store -r"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "3fd06d33-7a80-48e3-abc9-dec0a6cfd09d",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"!pip install -U sagemaker"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "629a43ad",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
"from sagemaker.feature_store.feature_group import FeatureGroup\n",
"from sagemaker.sklearn.estimator import SKLearn\n",
"from sagemaker.sklearn.model import SKLearnModel\n",
"from time import gmtime, strftime\n",
"import boto3\n",
"import sys\n",
"import sagemaker\n",
"import json\n",
"import os\n",
"import pandas as pd\n",
"from sagemaker.model_metrics import ModelMetrics, MetricsSource\n",
"from sagemaker.analytics import ExperimentAnalytics\n",
"from sagemaker.tuner import IntegerParameter, ContinuousParameter, HyperparameterTuner\n",
"# SageMaker Experiments\n",
"from sagemaker.experiments.run import Run\n",
"from sagemaker.utils import unique_name_from_base\n",
"\n",
"from sagemaker.feature_store.feature_group import FeatureGroup\n",
"from sagemaker import image_uris\n",
"from sagemaker.inputs import TrainingInput\n",
"\n",
"from helper_library import *"
]
},
{
"cell_type": "markdown",
"id": "bf525dbb",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"**Session variables**"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "279da59d",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
"# Useful SageMaker variables\n",
"sagemaker_session = sagemaker.Session()\n",
"bucket = sagemaker_session.default_bucket()\n",
"role_arn= sagemaker.get_execution_role()\n",
"region = sagemaker_session.boto_region_name\n",
"s3_client = boto3.client('s3', region_name=region)\n",
"sagemaker_client = boto3.client('sagemaker')\n",
"\n",
"enable_local_mode_training = False\n",
"model_package_group_name = 'synthetic-housing-models'\n",
"model_name = 'xgboost-model'\n",
"\n",
"\n",
"fs_dir = os.path.join(os.getcwd(), 'data/fs_data')\n",
"os.makedirs(fs_dir, exist_ok=True)\n",
"\n",
"fs_train_dir = os.path.join(os.getcwd(), 'data/fs_data/train')\n",
"os.makedirs(fs_train_dir, exist_ok=True)\n",
"\n",
"fs_validation_dir = os.path.join(os.getcwd(), 'data/fs_data/validation')\n",
"os.makedirs(fs_validation_dir, exist_ok=True)\n",
"\n",
"experiment_name = unique_name_from_base('synthetic-housing-regression')"
]
},
{
"cell_type": "markdown",
"id": "b72d1641",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"## Recap\n",
"\n",
"So we've processed our data and now have training and validation sets available in Feature Store to be used for training. Since SageMaker training jobs expects the training data to be on s3, let's first add our feature store data to s3"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "dc32606d",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
"def save_fs_data_to_s3(fg_name, features_to_select, sm_session, file_name, local_path, bucket, bucket_prefix):\n",
" fs_group = FeatureGroup(name=fg_name, sagemaker_session=sm_session) \n",
" query = fs_group.athena_query()\n",
" table = query.table_name\n",
" query_string = f'SELECT {features_to_select} FROM \"sagemaker_featurestore\".\"{table}\" ORDER BY record_id'\n",
" query_results= 'sagemaker-featurestore'\n",
" output_location = f's3://{bucket}/{query_results}/query_results/'\n",
" query.run(query_string=query_string, output_location=output_location)\n",
" query.wait()\n",
" df = query.as_dataframe()\n",
" df.to_csv(local_path+'/'+file_name, index=False, header=False)\n",
" s3_client.upload_file(local_path+'/'+file_name, bucket, bucket_prefix+'/'+file_name)\n",
" dataset_uri_prefix = \"s3://\" + bucket + \"/\" + bucket_prefix\n",
" return dataset_uri_prefix\n",
"\n",
"train_data = save_fs_data_to_s3(\n",
" train_feature_group_name, \n",
" features_to_select, \n",
" sagemaker_session, \n",
" \"train.csv\", \n",
" fs_train_dir, \n",
" bucket, \n",
" s3_prefix+\"/data/fs_data/train\"\n",
")\n",
"val_data = save_fs_data_to_s3(\n",
" validation_feature_group_name, \n",
" features_to_select, \n",
" sagemaker_session, \n",
" \"validation.csv\", \n",
" fs_validation_dir, \n",
" bucket, \n",
" s3_prefix+\"/data/fs_data/validation\"\n",
")\n",
"train_data, val_data"
]
},
{
"cell_type": "markdown",
"id": "f6e2e28d",
"metadata": {},
"source": [
"Let's compare the dataset distribution of our original dataset and the one read from Feature Store."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "8da5adba",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# read original training data\n",
"df_train_orig = pd.read_csv(sm_processed_train_dir+'/train.csv', header=None)\n",
"df_train_orig.describe()"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "be7e54b5",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"# reading training data from Feature Store\n",
"df_train_fs = pd.read_csv(fs_train_dir+'/train.csv', header=None)\n",
"df_train_fs.describe()"
]
},
{
"cell_type": "markdown",
"id": "989eacb9",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"Great! Our dataset distribution seems intact!\n",
"\n",
"We are ready to train a SageMaker Scikit-Learn model with it! At this point, you could choose to train locally if you're still experimenting with small datasets; however, we're going to move on to training at scale using transient compute environments using SageMaker Training Jobs."
]
},
{
"cell_type": "markdown",
"id": "af016e38",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"## SageMaker Training"
]
},
{
"cell_type": "markdown",
"id": "f53ae83f",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"Now that we've prepared our training and test data, we can move on to use SageMaker's hosted training functionality - [SageMaker Training](https://docs.aws.amazon.com/sagemaker/latest/dg/train-model.html). Hosted training is preferred for doing actual training, especially large-scale, distributed training. Unlike training a model on a local computer or server, SageMaker hosted training will spin up a separate cluster of machines managed by SageMaker to train your model. Before starting hosted training, the data must be in S3, or an EFS or FSx for Lustre file system. We uploaded to S3 in the previous notebook, so we're good to go here."
]
},
{
"attachments": {},
"cell_type": "markdown",
"id": "856fee14",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"Let's go ahead and create a Random Forest Regressor model from the Scikit-Learn library."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "891e02b3-ff7d-48a8-a27d-8f658e26681e",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"from sagemaker.sklearn.estimator import SKLearn\n",
"\n",
"hyperparameters = {'max_depth': 8, 'n_jobs': 4, 'n_estimators': 80}\n",
"train_instance_type = 'ml.c5.xlarge'\n",
"inputs = {'train': train_data, 'validation': val_data}\n",
" \n",
"# Metrics to be captured from logs.\n",
"metric_definitions = [{'Name': 'r_squared',\n",
" 'Regex': 'r-squared: ([0-9\\\\.]+)'},\n",
" {'Name': 'mse',\n",
" 'Regex': 'MSE: ([0-9\\\\.]+)'}]\n",
"\n",
"estimator_parameters = {\n",
" 'entry_point': './pipeline_scripts/train_deploy_scikitlearn_randomforestregressor.py',\n",
" 'framework_version': '0.23-1',\n",
" 'py_version': 'py3',\n",
" 'instance_type': train_instance_type,\n",
" 'instance_count': 1,\n",
" 'hyperparameters': hyperparameters,\n",
" 'role': role_arn,\n",
" 'metric_definitions': metric_definitions,\n",
" 'base_job_name': 'randomforestregressor-model',\n",
" 'output_path': f's3://{bucket}/{s3_prefix}/',\n",
" 'image_scope': 'training'\n",
"}"
]
},
{
"attachments": {},
"cell_type": "markdown",
"id": "a4d2337e",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"Before we actually train the Scikit-Learn model, we'll want to make sure we track experiments."
]
},
{
"cell_type": "markdown",
"id": "4ccd3201-a6e2-4d8f-87bd-78bed93e6102",
"metadata": {},
"source": [
"[SageMaker Experiments](https://docs.aws.amazon.com/sagemaker/latest/dg/experiments.html) can track all the model training iterations. Experiments are a great way to organize your data science work. You can create experiments to organize all your model development work for:\n",
"\n",
"1. A business use case you are addressing (e.g. create experiment named \"customer churn prediction\"), or\n",
"2. A data science team that owns the experiment (e.g. create experiment named \"marketing analytics experiment\"), or\n",
"3. A specific data science and ML project. Think of it as a \"folder\" for organizing your \"files\"."
]
},
{
"cell_type": "markdown",
"id": "ec27bb1b-ee54-4654-b92c-0c91cd517236",
"metadata": {},
"source": [
"To both train our model and track experiments, we'll just need a few lines of code below."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "c41d802d",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"scrolled": true,
"tags": []
},
"outputs": [],
"source": [
"from IPython.core.display import display, HTML\n",
"\n",
"display(\n",
" HTML(\n",
" 'Review the Training Job After About 5 Minutes'.format(\n",
" region, experiment_name\n",
" )\n",
" )\n",
")\n",
"\n",
"with Run(experiment_name=experiment_name, run_name='12345-test') as run:\n",
" estimator = SKLearn(**estimator_parameters)\n",
" estimator.fit(inputs)"
]
},
{
"cell_type": "markdown",
"id": "e93fbfab",
"metadata": {
"pycharm": {
"name": "#%% md\n"
},
"tags": []
},
"source": [
" Now that training finished, we can navigate to the Experiments menu in SageMaker Studio and take a gander at our model's MSE. You can find **SageMaker Experiments** on the left menu under **Home**. "
]
},
{
"attachments": {},
"cell_type": "markdown",
"id": "a9ca430c",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"If we don't like the MSE, we could improve it by adjusting model hyperparameters. But instead of guessing what hyperparameters we should have, we can let SageMaker search the hyperparameter space in an intelligent way on our behalf."
]
},
{
"cell_type": "markdown",
"id": "143c8b41",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"## SageMaker Training with Automatic Model Tuning (HPO)"
]
},
{
"cell_type": "markdown",
"id": "678aa910",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"[Amazon SageMaker Automatic Model Tuning](https://docs.aws.amazon.com/sagemaker/latest/dg/automatic-model-tuning.html), also known as hyperparameter tuning/optimization, finds the best version of a model by running many training jobs on your dataset using the algorithm and ranges of hyperparameters that you specify. It then chooses the hyperparameter values that result in a model that performs the best, as measured by a metric that you choose.\n",
"\n",
"You can use SageMaker automatic model tuning with built-in algorithms, custom algorithms, and SageMaker pre-built containers for machine learning frameworks."
]
},
{
"cell_type": "markdown",
"id": "efe599dd",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"We begin by specifying the hyperparameters we wish to tune, and the range of values over which to tune each one. We also must specify an objective metric to be optimized: in this use case, we'd like to minimize the validation loss."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "32bd7d44",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
"hyperparameter_ranges = {\n",
" 'max_depth': IntegerParameter(6, 10),\n",
" 'n_jobs': IntegerParameter(4, 6),\n",
" 'n_estimators': IntegerParameter(80, 130),\n",
"}\n",
"\n",
"objective_metric_name = 'mse'\n",
"objective_type = 'Minimize'"
]
},
{
"cell_type": "markdown",
"id": "5efce7e0",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"Next we specify a HyperparameterTuner object that takes the above definitions as parameters. Each tuning job must be given a budget: a maximum number of training jobs. A tuning job will complete after that many training jobs have been executed. \n",
"\n",
"We also can specify how much parallelism to employ, in this case two jobs, meaning that the tuning job will complete after two series of two jobs in parallel have completed (so, a total of 4 jobs as set by `max_jobs`). For the default Bayesian Optimization tuning strategy used here, the tuning search is informed by the results of previous groups of training jobs, so we don't run all of the jobs in parallel, but rather divide the jobs into groups of parallel jobs. There is a trade-off: using more parallel jobs will finish tuning sooner, but likely will sacrifice tuning search accuracy. \n",
"\n",
"Now we can launch a hyperparameter tuning job by calling the `fit` method of the HyperparameterTuner object. The tuning job may take some minutes to finish. While you're waiting, the status of the tuning job, including metadata and results for invidual training jobs within the tuning job, can be checked in the SageMaker console in the **Hyperparameter tuning jobs** panel. "
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "6b7a43ac",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
" tuner_parameters = {'estimator': estimator,\n",
" 'objective_metric_name': objective_metric_name,\n",
" 'hyperparameter_ranges': hyperparameter_ranges,\n",
" 'metric_definitions': metric_definitions,\n",
" 'max_jobs': 4,\n",
" 'max_parallel_jobs': 2,\n",
" 'objective_type': objective_type}\n",
" \n",
"tuner = HyperparameterTuner(**tuner_parameters)\n",
"\n",
"tuning_job_name = f'rf-model-tuning-{strftime(\"%d-%H-%M-%S\", gmtime())}'\n",
"display(\n",
" HTML(\n",
" 'Review the Tuning Job After About 5 Minutes'.format(\n",
" region, tuning_job_name\n",
" )\n",
" )\n",
")\n",
"tuner.fit(inputs, job_name=tuning_job_name)\n",
"tuner.wait()"
]
},
{
"cell_type": "markdown",
"id": "30750505",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"After the tuning job is finished, we can use the `HyperparameterTuningJobAnalytics` object from the SageMaker Python SDK to list the top 5 tuning jobs with the best performance. Although the results vary from tuning job to tuning job, the best validation loss from the tuning job (under the FinalObjectiveValue column) likely will be substantially lower than the validation loss from the hosted training job above, where we did not perform any tuning other than manually increasing the number of epochs once. "
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "12865b0e",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
"tuner_metrics = sagemaker.HyperparameterTuningJobAnalytics(tuning_job_name)\n",
"tuner_metrics.dataframe().sort_values(['FinalObjectiveValue'], ascending=True).head(5)"
]
},
{
"cell_type": "markdown",
"id": "572486b9",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"## Model Registry"
]
},
{
"cell_type": "markdown",
"id": "608dda7f",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"With the [SageMaker Model Registry](https://docs.aws.amazon.com/sagemaker/latest/dg/model-registry.html) you can do the following:\n",
"\n",
"- Catalog models for production.\n",
"- Manage model versions.\n",
"- Associate metadata, such as training metrics, with a model.\n",
"- Manage the approval status of a model.\n",
"- Deploy models to production.\n",
"- Automate model deployment with CI/CD.\n",
"\n",
"You can catalog models by creating model package groups that contain different versions of a model. You can create a model group that tracks all of the models that you train to solve a particular problem. You can then register each model you train and the model registry adds it to the model group as a new model version. A typical workflow might look like the following:\n",
"\n",
"- Create a model group.\n",
"- Create an ML pipeline that trains a model.\n",
"- For each run of the ML pipeline, create a model version that you register in the model group you created in the first step."
]
},
{
"cell_type": "markdown",
"id": "aebd02ed",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"So first we'll create a [Model Package Group](https://docs.aws.amazon.com/sagemaker/latest/dg/model-registry-model-group.html) in which we can store/group all related models and their versions."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "10d7611b",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
"sagemaker_client.create_model_package_group(ModelPackageGroupName=model_package_group_name,\n",
" ModelPackageGroupDescription='Models predicting synthetic housing prices') "
]
},
{
"cell_type": "markdown",
"id": "7de0cd93",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"Next we'll register the model we just trained with SageMaker Training."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "6911dab0",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
"# Register model\n",
"best_estimator = tuner.best_estimator()\n",
"model_metrics = create_training_job_metrics(best_estimator, s3_prefix, region, bucket)"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "2789f700-a57e-46cd-83af-b5c6dd3947e9",
"metadata": {
"tags": []
},
"outputs": [],
"source": [
"model_package = best_estimator.register(content_types=['text/csv'],\n",
" response_types=['application/json'],\n",
" inference_instances=['ml.t2.medium', 'ml.m5.xlarge'],\n",
" transform_instances=['ml.m5.xlarge'],\n",
" image_uri=best_estimator.image_uri,\n",
" model_package_group_name=model_package_group_name,\n",
" #model_metrics=model_metrics,\n",
" approval_status='PendingManualApproval',\n",
" description='XGBoost model to predict synthetic housing prices',\n",
" model_name=model_name,\n",
" name=model_name)\n",
"model_package_arn = model_package.model_package_arn"
]
},
{
"cell_type": "markdown",
"id": "1d60b072",
"metadata": {
"pycharm": {
"name": "#%% md\n"
}
},
"source": [
"We'll store relevant variables to be used in the next notebooks."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "d1ef883c",
"metadata": {
"pycharm": {
"name": "#%%\n"
},
"tags": []
},
"outputs": [],
"source": [
"%store model_package_arn\n",
"%store model_name\n",
"%store model_package_group_name\n",
"#%store model_metrics"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "d673611d-81be-4fa6-b162-81899d1079b0",
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"instance_type": "ml.t3.medium",
"kernelspec": {
"display_name": "Python 3 (Data Science)",
"language": "python",
"name": "python3__SAGEMAKER_INTERNAL__arn:aws:sagemaker:us-east-1:081325390199:image/datascience-1.0"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.10"
}
},
"nbformat": 4,
"nbformat_minor": 5
}